
slideflow
Powerful, open-source AI tools for digital pathology.
Stars: 260

Slideflow is a deep learning library for digital pathology, offering a user-friendly interface for model development. It is designed for medical researchers and AI enthusiasts, providing an accessible platform for developing state-of-the-art pathology models. Slideflow offers customizable training pipelines, robust slide processing and stain normalization toolkit, support for weakly-supervised or strongly-supervised labels, built-in foundation models, multiple-instance learning, self-supervised learning, generative adversarial networks, explainability tools, layer activation analysis tools, uncertainty quantification, interactive user interface for model deployment, and more. It supports both PyTorch and Tensorflow, with optional support for Libvips for slide reading. Slideflow can be installed via pip, Docker container, or from source, and includes non-commercial add-ons for additional tools and pretrained models. It allows users to create projects, extract tiles from slides, train models, and provides evaluation tools like heatmaps and mosaic maps.
README:
Slideflow is a deep learning library for digital pathology, offering a user-friendly interface for model development.
Designed for both medical researchers and AI enthusiasts, the goal of Slideflow is to provide an accessible, easy-to-use interface for developing state-of-the-art pathology models. Slideflow has been built with the future in mind, offering a scalable platform for digital biomarker development that bridges the gap between ever-evolving, sophisticated methods and the needs of a clinical researcher. For developers, Slideflow provides multiple endpoints for integration with other packages and external training paradigms, allowing you to leverage highly optimized, pathology-specific processes with the latest ML methodologies.
- Easy-to-use, highly customizable training pipelines
- Robust slide processing and stain normalization toolkit
- Support for training with weakly-supervised or strongly-supervised labels
- Built-in, state-of-the-art foundation models
- Multiple-instance learning (MIL)
- Self-supervised learning (SSL)
- Generative adversarial networks (GANs)
- Explainability tools: Heatmaps, mosaic maps, saliency maps, synthetic histology
- Robust layer activation analysis tools
- Uncertainty quantification
- Interactive user interface for model deployment
- ... and more!
Full documentation with example tutorials can be found at slideflow.dev.
- Python >= 3.7 (<3.10 if using cuCIM)
- PyTorch >= 1.9 or Tensorflow 2.5-2.11
- Libvips >= 8.9 (alternative slide reader, adds support for *.scn, *.mrxs, *.ndpi, *.vms, and *.vmu files).
- Linear solver (for preserved-site cross-validation)
- CPLEX 20.1.0 with Python API
- or Pyomo with Bonmin solver
Slideflow can be installed with PyPI, as a Docker container, or run from source.
pip3 install --upgrade setuptools pip wheel
pip3 install slideflow[cucim] cupy-cuda11x
The cupy package name depends on the installed CUDA version; see here for installation instructions. cupy is not required if using Libvips.
Alternatively, pre-configured docker images are available with OpenSlide/Libvips and the latest version of either Tensorflow and PyTorch. To install with the Tensorflow backend:
docker pull jamesdolezal/slideflow:latest-tf
docker run -it --gpus all jamesdolezal/slideflow:latest-tf
To install with the PyTorch backend:
docker pull jamesdolezal/slideflow:latest-torch
docker run -it --shm-size=2g --gpus all jamesdolezal/slideflow:latest-torch
To run from source, clone this repository, install the conda development environment, and build a wheel:
git clone https://github.com/slideflow/slideflow
conda env create -f slideflow/environment.yml
conda activate slideflow
pip install -e slideflow/ cupy-cuda11x
To add additional tools and pretrained models available under a non-commercial license, install slideflow-gpl and slideflow-noncommercial:
pip install slideflow-gpl slideflow-noncommercial
This will provide integrated access to 6 additional pretrained foundation models (UNI, HistoSSL, GigaPath, PLIP, RetCCL, and CTransPath), the MIL architecture CLAM, the UQ algorithm BISCUIT, and the GAN framework StyleGAN3.
Slideflow supports both PyTorch and Tensorflow, defaulting to PyTorch if both are available. You can specify the backend to use with the environmental variable SF_BACKEND. For example:
export SF_BACKEND=tensorflow
By default, Slideflow reads whole-slide images using cuCIM. Although much faster than other openslide-based frameworks, it supports fewer slide scanner formats. Slideflow also includes a Libvips backend, which adds support for *.scn, *.mrxs, *.ndpi, *.vms, and *.vmu files. You can set the active slide backend with the environmental variable SF_SLIDE_BACKEND:
export SF_SLIDE_BACKEND=libvips
Slideflow experiments are organized into Projects, which supervise storage of whole-slide images, extracted tiles, and patient-level annotations. The fastest way to get started is to use one of our preconfigured projects, which will automatically download slides from the Genomic Data Commons:
import slideflow as sf
P = sf.create_project(
root='/project/destination',
cfg=sf.project.LungAdenoSquam(),
download=True
)After the slides have been downloaded and verified, you can skip to Extract tiles from slides.
Alternatively, to create a new custom project, supply the location of patient-level annotations (CSV), slides, and a destination for TFRecords to be saved:
import slideflow as sf
P = sf.create_project(
'/project/path',
annotations="/patient/annotations.csv",
slides="/slides/directory",
tfrecords="/tfrecords/directory"
)Ensure that the annotations file has a slide column for each annotation entry with the filename (without extension) of the corresponding slide.
Next, whole-slide images are segmented into smaller image tiles and saved in *.tfrecords format. Extract tiles from slides at a given magnification (width in microns size) and resolution (width in pixels) using sf.Project.extract_tiles():
P.extract_tiles(
tile_px=299, # Tile size, in pixels
tile_um=302 # Tile size, in microns
)If slides are on a network drive or a spinning HDD, tile extraction can be accelerated by buffering slides to a SSD or ramdisk:
P.extract_tiles(
...,
buffer="/mnt/ramdisk"
)Once tiles are extracted, models can be trained. Start by configuring a set of hyperparameters:
params = sf.ModelParams(
tile_px=299,
tile_um=302,
batch_size=32,
model='xception',
learning_rate=0.0001,
...
)Models can then be trained using these parameters. Models can be trained to categorical, multi-categorical, continuous, or time-series outcomes, and the training process is highly configurable. For example, to train models in cross-validation to predict the outcome 'category1' as stored in the project annotations file:
P.train(
'category1',
params=params,
save_predictions=True,
multi_gpu=True
)Slideflow includes a host of additional tools, including model evaluation and prediction, heatmaps, analysis of layer activations, mosaic maps, and more. See our full documentation for more details and tutorials.
Slideflow has been used by:
- Dolezal et al, Modern Pathology, 2020
- Rosenberg et al, Journal of Clinical Oncology [abstract], 2020
- Howard et al, Nature Communications, 2021
- Dolezal et al Nature Communications, 2022
- Storozuk et al, Modern Pathology [abstract], 2022
- Partin et al Front Med, 2022
- Dolezal et al Journal of Clinical Oncology [abstract], 2022
- Dolezal et al Mediastinum [abstract], 2022
- Howard et al npj Breast Cancer, 2023
- Dolezal et al npj Precision Oncology, 2023
- Hieromnimon et al [bioRxiv], 2023
- Carrillo-Perez et al Cancer Imaging, 2023
This code is made available under the Apache-2.0 license.
If you find our work useful for your research, or if you use parts of this code, please consider citing as follows:
Dolezal, J.M., Kochanny, S., Dyer, E. et al. Slideflow: deep learning for digital histopathology with real-time whole-slide visualization. BMC Bioinformatics 25, 134 (2024). https://doi.org/10.1186/s12859-024-05758-x
@Article{Dolezal2024,
author={Dolezal, James M. and Kochanny, Sara and Dyer, Emma and Ramesh, Siddhi and Srisuwananukorn, Andrew and Sacco, Matteo and Howard, Frederick M. and Li, Anran and Mohan, Prajval and Pearson, Alexander T.},
title={Slideflow: deep learning for digital histopathology with real-time whole-slide visualization},
journal={BMC Bioinformatics},
year={2024},
month={Mar},
day={27},
volume={25},
number={1},
pages={134},
doi={10.1186/s12859-024-05758-x},
url={https://doi.org/10.1186/s12859-024-05758-x}
}
For Tasks:
Click tags to check more tools for each tasksFor Jobs:
Alternative AI tools for slideflow
Similar Open Source Tools

slideflow
Slideflow is a deep learning library for digital pathology, offering a user-friendly interface for model development. It is designed for medical researchers and AI enthusiasts, providing an accessible platform for developing state-of-the-art pathology models. Slideflow offers customizable training pipelines, robust slide processing and stain normalization toolkit, support for weakly-supervised or strongly-supervised labels, built-in foundation models, multiple-instance learning, self-supervised learning, generative adversarial networks, explainability tools, layer activation analysis tools, uncertainty quantification, interactive user interface for model deployment, and more. It supports both PyTorch and Tensorflow, with optional support for Libvips for slide reading. Slideflow can be installed via pip, Docker container, or from source, and includes non-commercial add-ons for additional tools and pretrained models. It allows users to create projects, extract tiles from slides, train models, and provides evaluation tools like heatmaps and mosaic maps.
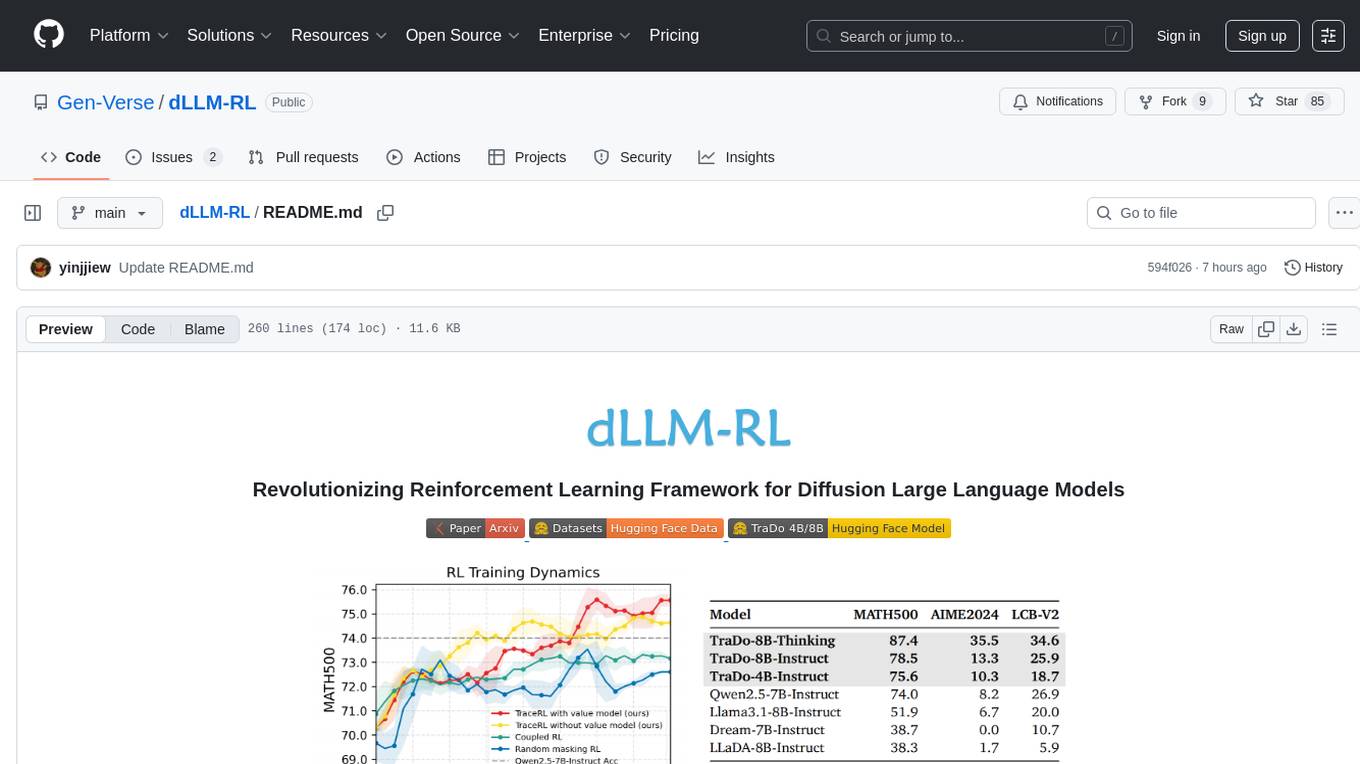
dLLM-RL
dLLM-RL is a revolutionary reinforcement learning framework designed for Diffusion Large Language Models. It supports various models with diverse structures, offers inference acceleration, RL training capabilities, and SFT functionalities. The tool introduces TraceRL for trajectory-aware RL and diffusion-based value models for optimization stability. Users can download and try models like TraDo-4B-Instruct and TraDo-8B-Instruct. The tool also provides support for multi-node setups and easy building of reinforcement learning methods. Additionally, it offers supervised fine-tuning strategies for different models and tasks.
labo
LABO is a time series forecasting and analysis framework that integrates pre-trained and fine-tuned LLMs with multi-domain agent-based systems. It allows users to create and tune agents easily for various scenarios, such as stock market trend prediction and web public opinion analysis. LABO requires a specific runtime environment setup, including system requirements, Python environment, dependency installations, and configurations. Users can fine-tune their own models using LABO's Low-Rank Adaptation (LoRA) for computational efficiency and continuous model updates. Additionally, LABO provides a Python library for building model training pipelines and customizing agents for specific tasks.
Genesis
Genesis is a physics platform designed for general purpose Robotics/Embodied AI/Physical AI applications. It includes a universal physics engine, a lightweight, ultra-fast, pythonic, and user-friendly robotics simulation platform, a powerful and fast photo-realistic rendering system, and a generative data engine that transforms user-prompted natural language description into various modalities of data. It aims to lower the barrier to using physics simulations, unify state-of-the-art physics solvers, and minimize human effort in collecting and generating data for robotics and other domains.
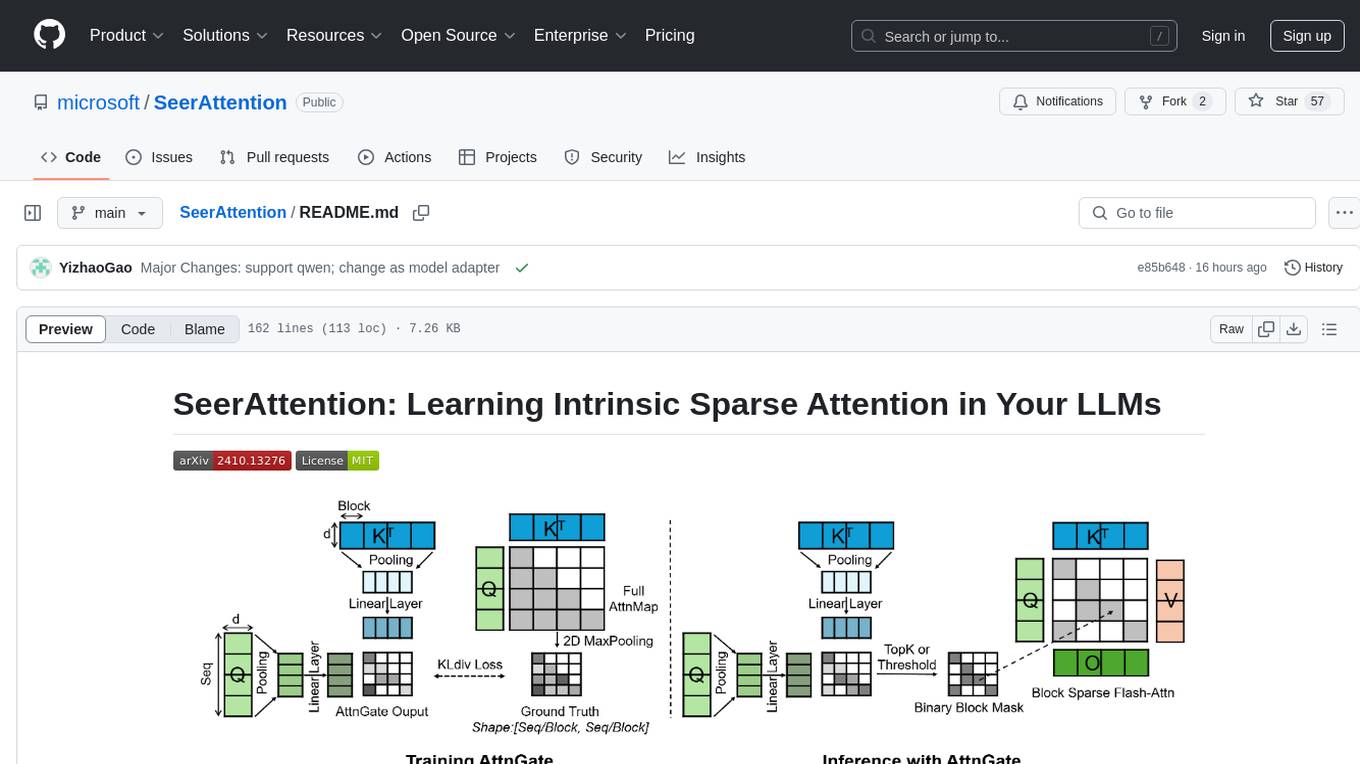
SeerAttention
SeerAttention is a novel trainable sparse attention mechanism that learns intrinsic sparsity patterns directly from LLMs through self-distillation at post-training time. It achieves faster inference while maintaining accuracy for long-context prefilling. The tool offers features such as trainable sparse attention, block-level sparsity, self-distillation, efficient kernel, and easy integration with existing transformer architectures. Users can quickly start using SeerAttention for inference with AttnGate Adapter and training attention gates with self-distillation. The tool provides efficient evaluation methods and encourages contributions from the community.
uccl
UCCL is a command-line utility tool designed to simplify the process of converting Unix-style file paths to Windows-style file paths and vice versa. It provides a convenient way for developers and system administrators to handle file path conversions without the need for manual adjustments. With UCCL, users can easily convert file paths between different operating systems, making it a valuable tool for cross-platform development and file management tasks.

only_train_once
Only Train Once (OTO) is an automatic, architecture-agnostic DNN training and compression framework that allows users to train a general DNN from scratch or a pretrained checkpoint to achieve high performance and slimmer architecture simultaneously in a one-shot manner without fine-tuning. The framework includes features for automatic structured pruning and erasing operators, as well as hybrid structured sparse optimizers for efficient model compression. OTO provides tools for pruning zero-invariant group partitioning, constructing pruned models, and visualizing pruning and erasing dependency graphs. It supports the HESSO optimizer and offers a sanity check for compliance testing on various DNNs. The repository also includes publications, installation instructions, quick start guides, and a roadmap for future enhancements and collaborations.
habitat-lab
Habitat-Lab is a modular high-level library for end-to-end development in embodied AI. It is designed to train agents to perform a wide variety of embodied AI tasks in indoor environments, as well as develop agents that can interact with humans in performing these tasks.
MMC
This repository, MMC, focuses on advancing multimodal chart understanding through large-scale instruction tuning. It introduces a dataset supporting various tasks and chart types, a benchmark for evaluating reasoning capabilities over charts, and an assistant achieving state-of-the-art performance on chart QA benchmarks. The repository provides data for chart-text alignment, benchmarking, and instruction tuning, along with existing datasets used in experiments. Additionally, it offers a Gradio demo for the MMCA model.
FATE-LLM
FATE-LLM is a framework supporting federated learning for large and small language models. It promotes training efficiency of federated LLMs using Parameter-Efficient methods, protects the IP of LLMs using FedIPR, and ensures data privacy during training and inference through privacy-preserving mechanisms.

MineStudio
MineStudio is a simple and efficient Minecraft development kit for AI research. It contains tools and APIs for developing Minecraft AI agents, including a customizable simulator, trajectory data structure, policy models, offline and online training pipelines, inference framework, and benchmarking automation. The repository is under development and welcomes contributions and suggestions.

HolmesVAD
Holmes-VAD is a framework for unbiased and explainable Video Anomaly Detection using multimodal instructions. It addresses biased detection in challenging events by leveraging precise temporal supervision and rich multimodal instructions. The framework includes a largescale VAD instruction-tuning benchmark, VAD-Instruct50k, created with single-frame annotations and a robust video captioner. It offers accurate anomaly localization and comprehensive explanations through a customized solution for interpretable video anomaly detection.
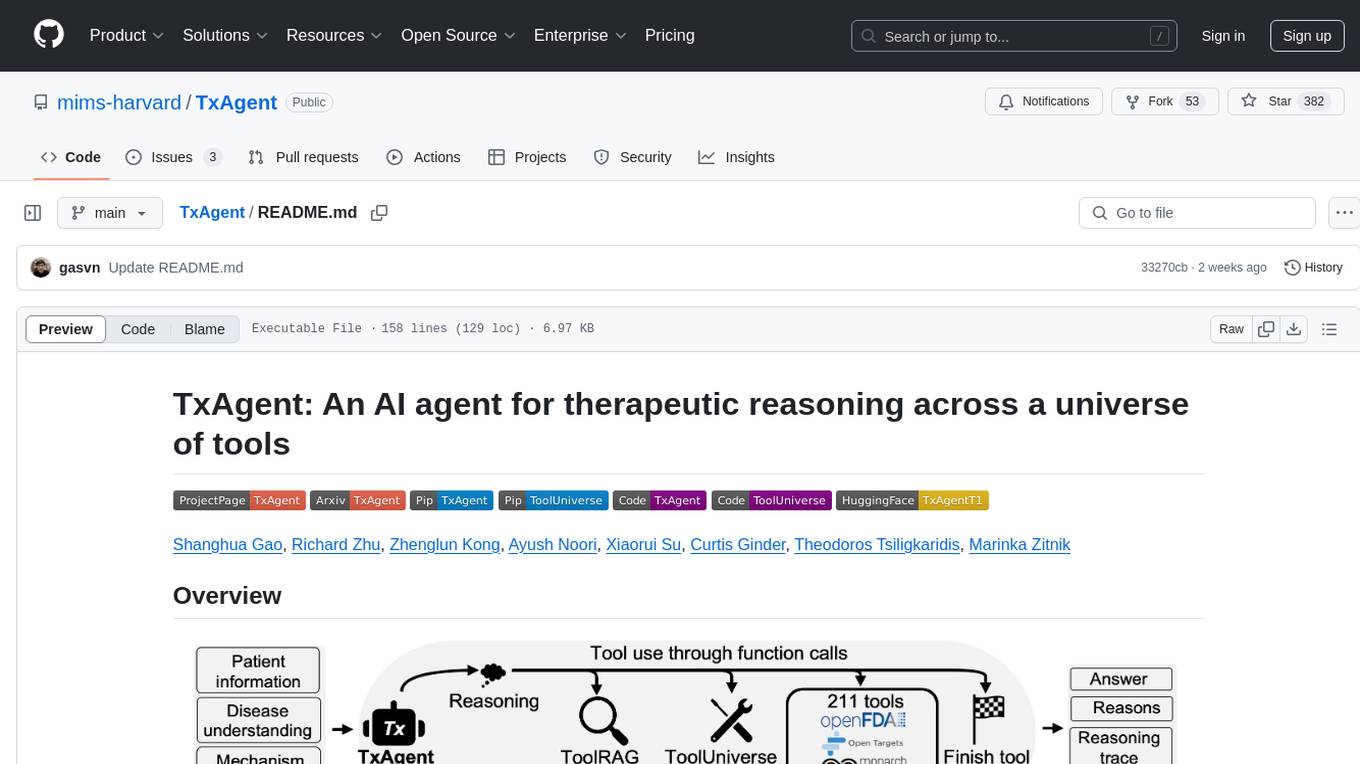
TxAgent
TxAgent is an AI agent designed for precision therapeutics, leveraging multi-step reasoning and real-time biomedical knowledge retrieval across a toolbox of 211 tools. It evaluates drug interactions, contraindications, and tailors treatment strategies to individual patient characteristics. TxAgent outperforms leading models across various drug reasoning tasks and personalized treatment scenarios, ensuring treatment recommendations align with clinical guidelines and real-world evidence.
ALMA
ALMA (Advanced Language Model-based Translator) is a many-to-many LLM-based translation model that utilizes a two-step fine-tuning process on monolingual and parallel data to achieve strong translation performance. ALMA-R builds upon ALMA models with LoRA fine-tuning and Contrastive Preference Optimization (CPO) for even better performance, surpassing GPT-4 and WMT winners. The repository provides ALMA and ALMA-R models, datasets, environment setup, evaluation scripts, training guides, and data information for users to leverage these models for translation tasks.

catalyst
Catalyst is a C# Natural Language Processing library designed for speed, inspired by spaCy's design. It provides pre-trained models, support for training word and document embeddings, and flexible entity recognition models. The library is fast, modern, and pure-C#, supporting .NET standard 2.0. It is cross-platform, running on Windows, Linux, macOS, and ARM. Catalyst offers non-destructive tokenization, named entity recognition, part-of-speech tagging, language detection, and efficient binary serialization. It includes pre-built models for language packages and lemmatization. Users can store and load models using streams. Getting started with Catalyst involves installing its NuGet Package and setting the storage to use the online repository. The library supports lazy loading of models from disk or online. Users can take advantage of C# lazy evaluation and native multi-threading support to process documents in parallel. Training a new FastText word2vec embedding model is straightforward, and Catalyst also provides algorithms for fast embedding search and dimensionality reduction.

raga-llm-hub
Raga LLM Hub is a comprehensive evaluation toolkit for Language and Learning Models (LLMs) with over 100 meticulously designed metrics. It allows developers and organizations to evaluate and compare LLMs effectively, establishing guardrails for LLMs and Retrieval Augmented Generation (RAG) applications. The platform assesses aspects like Relevance & Understanding, Content Quality, Hallucination, Safety & Bias, Context Relevance, Guardrails, and Vulnerability scanning, along with Metric-Based Tests for quantitative analysis. It helps teams identify and fix issues throughout the LLM lifecycle, revolutionizing reliability and trustworthiness.
For similar tasks

slideflow
Slideflow is a deep learning library for digital pathology, offering a user-friendly interface for model development. It is designed for medical researchers and AI enthusiasts, providing an accessible platform for developing state-of-the-art pathology models. Slideflow offers customizable training pipelines, robust slide processing and stain normalization toolkit, support for weakly-supervised or strongly-supervised labels, built-in foundation models, multiple-instance learning, self-supervised learning, generative adversarial networks, explainability tools, layer activation analysis tools, uncertainty quantification, interactive user interface for model deployment, and more. It supports both PyTorch and Tensorflow, with optional support for Libvips for slide reading. Slideflow can be installed via pip, Docker container, or from source, and includes non-commercial add-ons for additional tools and pretrained models. It allows users to create projects, extract tiles from slides, train models, and provides evaluation tools like heatmaps and mosaic maps.
ai-on-gke
This repository contains assets related to AI/ML workloads on Google Kubernetes Engine (GKE). Run optimized AI/ML workloads with Google Kubernetes Engine (GKE) platform orchestration capabilities. A robust AI/ML platform considers the following layers: Infrastructure orchestration that support GPUs and TPUs for training and serving workloads at scale Flexible integration with distributed computing and data processing frameworks Support for multiple teams on the same infrastructure to maximize utilization of resources

ray
Ray is a unified framework for scaling AI and Python applications. It consists of a core distributed runtime and a set of AI libraries for simplifying ML compute, including Data, Train, Tune, RLlib, and Serve. Ray runs on any machine, cluster, cloud provider, and Kubernetes, and features a growing ecosystem of community integrations. With Ray, you can seamlessly scale the same code from a laptop to a cluster, making it easy to meet the compute-intensive demands of modern ML workloads.

labelbox-python
Labelbox is a data-centric AI platform for enterprises to develop, optimize, and use AI to solve problems and power new products and services. Enterprises use Labelbox to curate data, generate high-quality human feedback data for computer vision and LLMs, evaluate model performance, and automate tasks by combining AI and human-centric workflows. The academic & research community uses Labelbox for cutting-edge AI research.
djl
Deep Java Library (DJL) is an open-source, high-level, engine-agnostic Java framework for deep learning. It is designed to be easy to get started with and simple to use for Java developers. DJL provides a native Java development experience and allows users to integrate machine learning and deep learning models with their Java applications. The framework is deep learning engine agnostic, enabling users to switch engines at any point for optimal performance. DJL's ergonomic API interface guides users with best practices to accomplish deep learning tasks, such as running inference and training neural networks.
mlflow
MLflow is a platform to streamline machine learning development, including tracking experiments, packaging code into reproducible runs, and sharing and deploying models. MLflow offers a set of lightweight APIs that can be used with any existing machine learning application or library (TensorFlow, PyTorch, XGBoost, etc), wherever you currently run ML code (e.g. in notebooks, standalone applications or the cloud). MLflow's current components are:
* `MLflow Tracking
tt-metal
TT-NN is a python & C++ Neural Network OP library. It provides a low-level programming model, TT-Metalium, enabling kernel development for Tenstorrent hardware.
burn
Burn is a new comprehensive dynamic Deep Learning Framework built using Rust with extreme flexibility, compute efficiency and portability as its primary goals.
For similar jobs
grand-challenge.org
Grand Challenge is a platform that provides access to large amounts of annotated training data, objective comparisons of state-of-the-art machine learning solutions, and clinical validation using real-world data. It assists researchers, data scientists, and clinicians in collaborating to develop robust machine learning solutions to problems in biomedical imaging.
Detection-and-Classification-of-Alzheimers-Disease
This tool is designed to detect and classify Alzheimer's Disease using Deep Learning and Machine Learning algorithms on an early basis, which is further optimized using the Crow Search Algorithm (CSA). Alzheimer's is a fatal disease, and early detection is crucial for patients to predetermine their condition and prevent its progression. By analyzing MRI scanned images using Artificial Intelligence technology, this tool can classify patients who may or may not develop AD in the future. The CSA algorithm, combined with ML algorithms, has proven to be the most effective approach for this purpose.

OpenCRISPR
OpenCRISPR is a set of free and open gene editing systems designed by Profluent Bio. The OpenCRISPR-1 protein maintains the prototypical architecture of a Type II Cas9 nuclease but is hundreds of mutations away from SpCas9 or any other known natural CRISPR-associated protein. You can view OpenCRISPR-1 as a drop-in replacement for many protocols that need a cas9-like protein with an NGG PAM and you can even use it with canonical SpCas9 gRNAs. OpenCRISPR-1 can be fused in a deactivated or nickase format for next generation gene editing techniques like base, prime, or epigenome editing.

AlphaFold3
AlphaFold3 is an implementation of the Alpha Fold 3 model in PyTorch for accurate structure prediction of biomolecular interactions. It includes modules for genetic diffusion and full model examples for forward pass computations. The tool allows users to generate random pair and single representations, operate on atomic coordinates, and perform structure predictions based on input tensors. The implementation also provides functionalities for training and evaluating the model.
fuse-med-ml
FuseMedML is a Python framework designed to accelerate machine learning-based discovery in the medical field by promoting code reuse. It provides a flexible design concept where data is stored in a nested dictionary, allowing easy handling of multi-modality information. The framework includes components for creating custom models, loss functions, metrics, and data processing operators. Additionally, FuseMedML offers 'batteries included' key components such as fuse.data for data processing, fuse.eval for model evaluation, and fuse.dl for reusable deep learning components. It supports PyTorch and PyTorch Lightning libraries and encourages the creation of domain extensions for specific medical domains.
hi-ml
The Microsoft Health Intelligence Machine Learning Toolbox is a repository that provides low-level and high-level building blocks for Machine Learning / AI researchers and practitioners. It simplifies and streamlines work on deep learning models for healthcare and life sciences by offering tested components such as data loaders, pre-processing tools, deep learning models, and cloud integration utilities. The repository includes two Python packages, 'hi-ml-azure' for helper functions in AzureML, 'hi-ml' for ML components, and 'hi-ml-cpath' for models and workflows related to histopathology images.
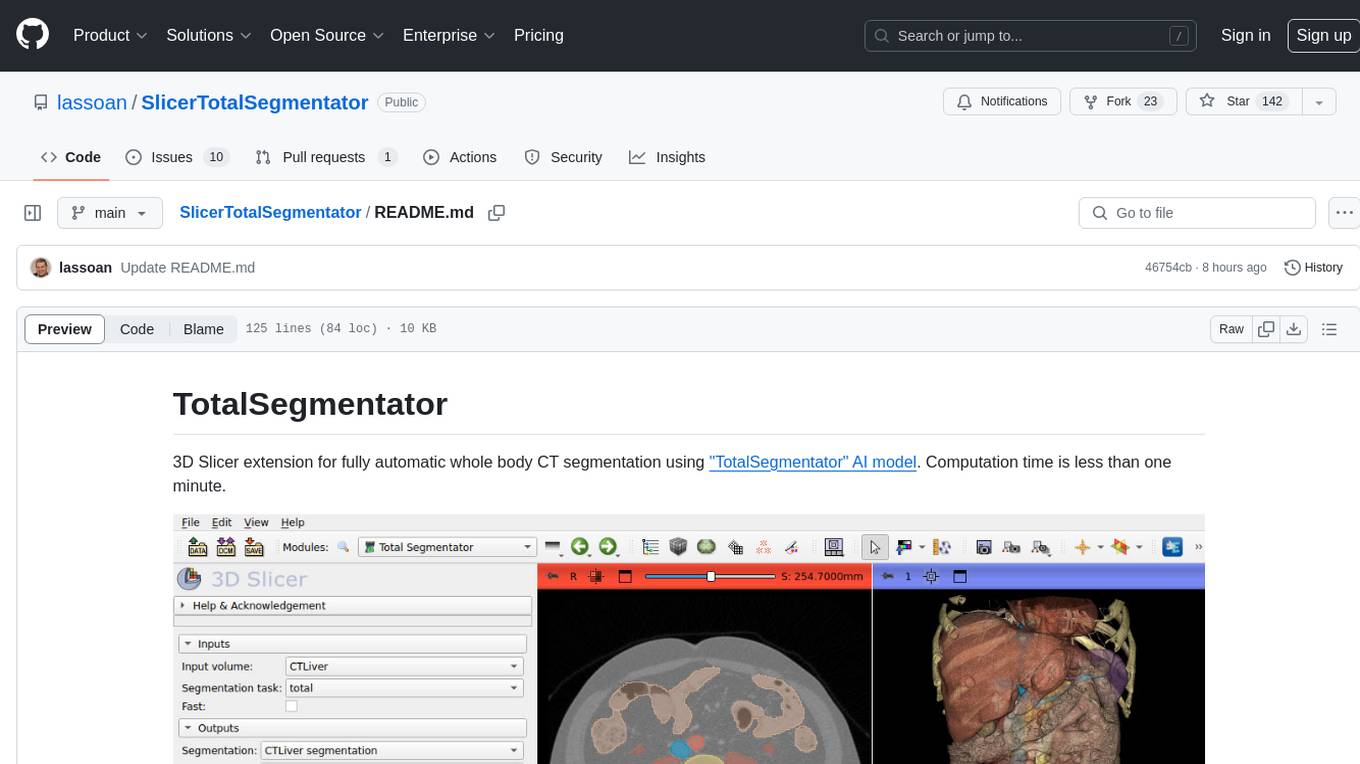
SlicerTotalSegmentator
TotalSegmentator is a 3D Slicer extension designed for fully automatic whole body CT segmentation using the 'TotalSegmentator' AI model. The computation time is less than one minute, making it efficient for research purposes. Users can set up GPU acceleration for faster segmentation. The tool provides a user-friendly interface for loading CT images, creating segmentations, and displaying results in 3D. Troubleshooting steps are available for common issues such as failed computation, GPU errors, and inaccurate segmentations. Contributions to the extension are welcome, following 3D Slicer contribution guidelines.
md-agent
MD-Agent is a LLM-agent based toolset for Molecular Dynamics. It uses Langchain and a collection of tools to set up and execute molecular dynamics simulations, particularly in OpenMM. The tool assists in environment setup, installation, and usage by providing detailed steps. It also requires API keys for certain functionalities, such as OpenAI and paper-qa for literature searches. Contributions to the project are welcome, with a detailed Contributor's Guide available for interested individuals.