
md-agent
Molecular dynamics simulations with an LLM agent
Stars: 73
MD-Agent is a LLM-agent based toolset for Molecular Dynamics. It uses Langchain and a collection of tools to set up and execute molecular dynamics simulations, particularly in OpenMM. The tool assists in environment setup, installation, and usage by providing detailed steps. It also requires API keys for certain functionalities, such as OpenAI and paper-qa for literature searches. Contributions to the project are welcome, with a detailed Contributor's Guide available for interested individuals.
README:
MDAgent is a LLM-agent based toolset for Molecular Dynamics. It's built using Langchain and uses a collection of tools to set up and execute molecular dynamics simulations, particularly in OpenMM.
To use the OpenMM features in the agent, please set up a conda environment, following these steps.
conda env create -n mdagent -f environment.yaml
conda activate mdagent
If you already have a conda environment, you can install dependencies before you activate it with the following step.
- Install the necessary conda dependencies:
conda env update -n <YOUR_CONDA_ENV_HERE> -f environment.yaml
pip install git+https://github.com/ur-whitelab/md-agent.git
The next step is to set up your API keys in your environment. An API key for LLM provider is necessary for this project. Supported LLM providers are OpenAI, TogetherAI, Fireworks, and Anthropic. Other tools require API keys, such as paper-qa for literature searches. We recommend setting up the keys in a .env file. You can use the provided .env.example file as a template.
- Copy the
.env.examplefile and rename it to.env:cp .env.example .env - Replace the placeholder values in
.envwith your actual keys
You can ask MDAgent to conduct molecular dynamics tasks using OpenAI's GPT model
from mdagent import MDAgent
agent = MDAgent(model="gpt-3.5-turbo")
agent.run("Simulate protein 1ZNI at 300 K for 0.1 ps and calculate the RMSD over time.")
Note: to distinguish Together models from the rest, you'll need to add "together" prefix in model flag, such as agent = MDAgent(model="together/meta-llama/Meta-Llama-3.1-70B-Instruct-Turbo")
By default, we support LLMs through OpenAI API. However, feel free to use other LLM providers. Make sure to install the necessary package for it. Here's list of packages required for alternative LLM providers we support:
-
pip install langchain-togetherto use models from TogetherAI -
pip install langchain-anthropicto use models from Anthropic -
pip install langchain-fireworksto use models from Fireworks
We welcome contributions to MDAgent! If you're interested in contributing to the project, please check out our Contributor's Guide for detailed instructions on getting started, feature development, and the pull request process.
We value and appreciate all contributions to MDAgent.
For Tasks:
Click tags to check more tools for each tasksFor Jobs:
Alternative AI tools for md-agent
Similar Open Source Tools

md-agent
MD-Agent is a LLM-agent based toolset for Molecular Dynamics. It uses Langchain and a collection of tools to set up and execute molecular dynamics simulations, particularly in OpenMM. The tool assists in environment setup, installation, and usage by providing detailed steps. It also requires API keys for certain functionalities, such as OpenAI and paper-qa for literature searches. Contributions to the project are welcome, with a detailed Contributor's Guide available for interested individuals.
atomic_agents
Atomic Agents is a modular and extensible framework designed for creating powerful applications. It follows the principles of Atomic Design, emphasizing small and single-purpose components. Leveraging Pydantic for data validation and serialization, the framework offers a set of tools and agents that can be combined to build AI applications. It depends on the Instructor package and supports various APIs like OpenAI, Cohere, Anthropic, and Gemini. Atomic Agents is suitable for developers looking to create AI agents with a focus on modularity and flexibility.

eureka-ml-insights
The Eureka ML Insights Framework is a repository containing code designed to help researchers and practitioners run reproducible evaluations of generative models efficiently. Users can define custom pipelines for data processing, inference, and evaluation, as well as utilize pre-defined evaluation pipelines for key benchmarks. The framework provides a structured approach to conducting experiments and analyzing model performance across various tasks and modalities.
lumigator
Lumigator is an open-source platform developed by Mozilla.ai to help users select the most suitable language model for their specific needs. It supports the evaluation of summarization tasks using sequence-to-sequence models such as BART and BERT, as well as causal models like GPT and Mistral. The platform aims to make model selection transparent, efficient, and empowering by providing a framework for comparing LLMs using task-specific metrics to evaluate how well a model fits a project's needs. Lumigator is in the early stages of development and plans to expand support to additional machine learning tasks and use cases in the future.
aisuite
Aisuite is a simple, unified interface to multiple Generative AI providers. It allows developers to easily interact with various Language Model (LLM) providers like OpenAI, Anthropic, Azure, Google, AWS, and more through a standardized interface. The library focuses on chat completions and provides a thin wrapper around python client libraries, enabling creators to test responses from different LLM providers without changing their code. Aisuite maximizes stability by using HTTP endpoints or SDKs for making calls to the providers. Users can install the base package or specific provider packages, set up API keys, and utilize the library to generate chat completion responses from different models.
engine-core
Engine Core is a project that demonstrates a pattern for enabling Large Language Models (LLMs) to undertake tasks with a dynamic system prompt and a collection of tool functions known as chat strategies. These strategies allow for the dynamic alteration of chat history, system prompts, and available tools on every run. The project includes example strategies such as demoStrategy, backendStrategy, and shellStrategy. Additionally, LLM integrations like Anthropic or OpenAI have been extracted into adapters to enable running the same app code and strategies while switching foundation models.
PromptAgent
PromptAgent is a repository for a novel automatic prompt optimization method that crafts expert-level prompts using language models. It provides a principled framework for prompt optimization by unifying prompt sampling and rewarding using MCTS algorithm. The tool supports different models like openai, palm, and huggingface models. Users can run PromptAgent to optimize prompts for specific tasks by strategically sampling model errors, generating error feedbacks, simulating future rewards, and searching for high-reward paths leading to expert prompts.
BentoDiffusion
BentoDiffusion is a BentoML example project that demonstrates how to serve and deploy diffusion models in the Stable Diffusion (SD) family. These models are specialized in generating and manipulating images based on text prompts. The project provides a guide on using SDXL Turbo as an example, along with instructions on prerequisites, installing dependencies, running the BentoML service, and deploying to BentoCloud. Users can interact with the deployed service using Swagger UI or other methods. Additionally, the project offers the option to choose from various diffusion models available in the repository for deployment.
ModernBERT
ModernBERT is a repository focused on modernizing BERT through architecture changes and scaling. It introduces FlexBERT, a modular approach to encoder building blocks, and heavily relies on .yaml configuration files to build models. The codebase builds upon MosaicBERT and incorporates Flash Attention 2. The repository is used for pre-training and GLUE evaluations, with a focus on reproducibility and documentation. It provides a collaboration between Answer.AI, LightOn, and friends.
sublayer
Sublayer is a model-agnostic Ruby AI Agent framework that provides base classes for building Generators, Actions, Tasks, and Agents to create AI-powered applications in Ruby. It supports various AI models and providers, such as OpenAI, Gemini, and Claude. Generators generate specific outputs, Actions perform operations, Agents are autonomous entities for tasks or monitoring, and Triggers decide when Agents are activated. The framework offers sample Generators and usage examples for building AI applications.
GhostOS
GhostOS is an AI Agent framework designed to replace JSON Schema with a Turing-complete code interaction interface (Moss Protocol). It aims to create intelligent entities capable of continuous learning and growth through code generation and project management. The framework supports various capabilities such as turning Python files into web agents, real-time voice conversation, body movements control, and emotion expression. GhostOS is still in early experimental development and focuses on out-of-the-box capabilities for AI agents.
council
Council is an open-source platform designed for the rapid development and deployment of customized generative AI applications using teams of agents. It extends the LLM tool ecosystem by providing advanced control flow and scalable oversight for AI agents. Users can create sophisticated agents with predictable behavior by leveraging Council's powerful approach to control flow using Controllers, Filters, Evaluators, and Budgets. The framework allows for automated routing between agents, comparing, evaluating, and selecting the best results for a task. Council aims to facilitate packaging and deploying agents at scale on multiple platforms while enabling enterprise-grade monitoring and quality control.
wandb
Weights & Biases (W&B) is a platform that helps users build better machine learning models faster by tracking and visualizing all components of the machine learning pipeline, from datasets to production models. It offers tools for tracking, debugging, evaluating, and monitoring machine learning applications. W&B provides integrations with popular frameworks like PyTorch, TensorFlow/Keras, Hugging Face Transformers, PyTorch Lightning, XGBoost, and Sci-Kit Learn. Users can easily log metrics, visualize performance, and compare experiments using W&B. The platform also supports hosting options in the cloud or on private infrastructure, making it versatile for various deployment needs.
quick-start-connectors
Cohere's Build-Your-Own-Connector framework allows integration of Cohere's Command LLM via the Chat API endpoint to any datastore/software holding text information with a search endpoint. Enables user queries grounded in proprietary information. Use-cases include question/answering, knowledge working, comms summary, and research. Repository provides code for popular datastores and a template connector. Requires Python 3.11+ and Poetry. Connectors can be built and deployed using Docker. Environment variables set authorization values. Pre-commits for linting. Connectors tailored to integrate with Cohere's Chat API for creating chatbots. Connectors return documents as JSON objects for Cohere's API to generate answers with citations.
REINVENT4
REINVENT is a molecular design tool for de novo design, scaffold hopping, R-group replacement, linker design, molecule optimization, and other small molecule design tasks. It uses a Reinforcement Learning (RL) algorithm to generate optimized molecules compliant with a user-defined property profile defined as a multi-component score. Transfer Learning (TL) can be used to create or pre-train a model that generates molecules closer to a set of input molecules.
inspect_ai
Inspect AI is a framework developed by the UK AI Safety Institute for evaluating large language models. It offers various built-in components for prompt engineering, tool usage, multi-turn dialog, and model graded evaluations. Users can extend Inspect by adding new elicitation and scoring techniques through additional Python packages. The tool aims to provide a comprehensive solution for assessing the performance and safety of language models.
For similar tasks
Azure-Analytics-and-AI-Engagement
The Azure-Analytics-and-AI-Engagement repository provides packaged Industry Scenario DREAM Demos with ARM templates (Containing a demo web application, Power BI reports, Synapse resources, AML Notebooks etc.) that can be deployed in a customer’s subscription using the CAPE tool within a matter of few hours. Partners can also deploy DREAM Demos in their own subscriptions using DPoC.
sorrentum
Sorrentum is an open-source project that aims to combine open-source development, startups, and brilliant students to build machine learning, AI, and Web3 / DeFi protocols geared towards finance and economics. The project provides opportunities for internships, research assistantships, and development grants, as well as the chance to work on cutting-edge problems, learn about startups, write academic papers, and get internships and full-time positions at companies working on Sorrentum applications.

tidb
TiDB is an open-source distributed SQL database that supports Hybrid Transactional and Analytical Processing (HTAP) workloads. It is MySQL compatible and features horizontal scalability, strong consistency, and high availability.

zep-python
Zep is an open-source platform for building and deploying large language model (LLM) applications. It provides a suite of tools and services that make it easy to integrate LLMs into your applications, including chat history memory, embedding, vector search, and data enrichment. Zep is designed to be scalable, reliable, and easy to use, making it a great choice for developers who want to build LLM-powered applications quickly and easily.
telemetry-airflow
This repository codifies the Airflow cluster that is deployed at workflow.telemetry.mozilla.org (behind SSO) and commonly referred to as "WTMO" or simply "Airflow". Some links relevant to users and developers of WTMO: * The `dags` directory in this repository contains some custom DAG definitions * Many of the DAGs registered with WTMO don't live in this repository, but are instead generated from ETL task definitions in bigquery-etl * The Data SRE team maintains a WTMO Developer Guide (behind SSO)

mojo
Mojo is a new programming language that bridges the gap between research and production by combining Python syntax and ecosystem with systems programming and metaprogramming features. Mojo is still young, but it is designed to become a superset of Python over time.
pandas-ai
PandasAI is a Python library that makes it easy to ask questions to your data in natural language. It helps you to explore, clean, and analyze your data using generative AI.
databend
Databend is an open-source cloud data warehouse that serves as a cost-effective alternative to Snowflake. With its focus on fast query execution and data ingestion, it's designed for complex analysis of the world's largest datasets.
For similar jobs
grand-challenge.org
Grand Challenge is a platform that provides access to large amounts of annotated training data, objective comparisons of state-of-the-art machine learning solutions, and clinical validation using real-world data. It assists researchers, data scientists, and clinicians in collaborating to develop robust machine learning solutions to problems in biomedical imaging.
Detection-and-Classification-of-Alzheimers-Disease
This tool is designed to detect and classify Alzheimer's Disease using Deep Learning and Machine Learning algorithms on an early basis, which is further optimized using the Crow Search Algorithm (CSA). Alzheimer's is a fatal disease, and early detection is crucial for patients to predetermine their condition and prevent its progression. By analyzing MRI scanned images using Artificial Intelligence technology, this tool can classify patients who may or may not develop AD in the future. The CSA algorithm, combined with ML algorithms, has proven to be the most effective approach for this purpose.

OpenCRISPR
OpenCRISPR is a set of free and open gene editing systems designed by Profluent Bio. The OpenCRISPR-1 protein maintains the prototypical architecture of a Type II Cas9 nuclease but is hundreds of mutations away from SpCas9 or any other known natural CRISPR-associated protein. You can view OpenCRISPR-1 as a drop-in replacement for many protocols that need a cas9-like protein with an NGG PAM and you can even use it with canonical SpCas9 gRNAs. OpenCRISPR-1 can be fused in a deactivated or nickase format for next generation gene editing techniques like base, prime, or epigenome editing.

AlphaFold3
AlphaFold3 is an implementation of the Alpha Fold 3 model in PyTorch for accurate structure prediction of biomolecular interactions. It includes modules for genetic diffusion and full model examples for forward pass computations. The tool allows users to generate random pair and single representations, operate on atomic coordinates, and perform structure predictions based on input tensors. The implementation also provides functionalities for training and evaluating the model.
fuse-med-ml
FuseMedML is a Python framework designed to accelerate machine learning-based discovery in the medical field by promoting code reuse. It provides a flexible design concept where data is stored in a nested dictionary, allowing easy handling of multi-modality information. The framework includes components for creating custom models, loss functions, metrics, and data processing operators. Additionally, FuseMedML offers 'batteries included' key components such as fuse.data for data processing, fuse.eval for model evaluation, and fuse.dl for reusable deep learning components. It supports PyTorch and PyTorch Lightning libraries and encourages the creation of domain extensions for specific medical domains.
hi-ml
The Microsoft Health Intelligence Machine Learning Toolbox is a repository that provides low-level and high-level building blocks for Machine Learning / AI researchers and practitioners. It simplifies and streamlines work on deep learning models for healthcare and life sciences by offering tested components such as data loaders, pre-processing tools, deep learning models, and cloud integration utilities. The repository includes two Python packages, 'hi-ml-azure' for helper functions in AzureML, 'hi-ml' for ML components, and 'hi-ml-cpath' for models and workflows related to histopathology images.
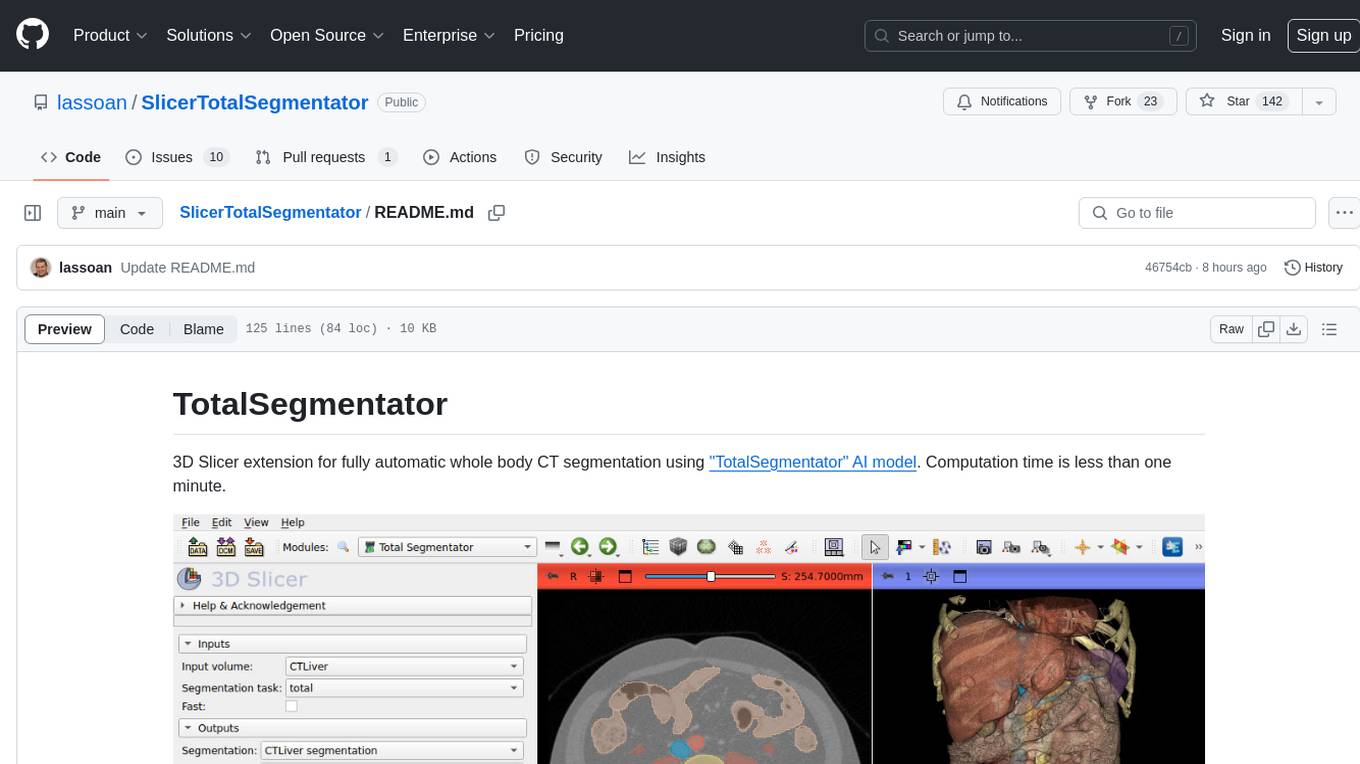
SlicerTotalSegmentator
TotalSegmentator is a 3D Slicer extension designed for fully automatic whole body CT segmentation using the 'TotalSegmentator' AI model. The computation time is less than one minute, making it efficient for research purposes. Users can set up GPU acceleration for faster segmentation. The tool provides a user-friendly interface for loading CT images, creating segmentations, and displaying results in 3D. Troubleshooting steps are available for common issues such as failed computation, GPU errors, and inaccurate segmentations. Contributions to the extension are welcome, following 3D Slicer contribution guidelines.

md-agent
MD-Agent is a LLM-agent based toolset for Molecular Dynamics. It uses Langchain and a collection of tools to set up and execute molecular dynamics simulations, particularly in OpenMM. The tool assists in environment setup, installation, and usage by providing detailed steps. It also requires API keys for certain functionalities, such as OpenAI and paper-qa for literature searches. Contributions to the project are welcome, with a detailed Contributor's Guide available for interested individuals.