
fuse-med-ml
A python framework accelerating ML based discovery in the medical field by encouraging code reuse. Batteries included :)
Stars: 138
FuseMedML is a Python framework designed to accelerate machine learning-based discovery in the medical field by promoting code reuse. It provides a flexible design concept where data is stored in a nested dictionary, allowing easy handling of multi-modality information. The framework includes components for creating custom models, loss functions, metrics, and data processing operators. Additionally, FuseMedML offers 'batteries included' key components such as fuse.data for data processing, fuse.eval for model evaluation, and fuse.dl for reusable deep learning components. It supports PyTorch and PyTorch Lightning libraries and encourages the creation of domain extensions for specific medical domains.
README:
A python framework accelerating ML based discovery in the medical field by encouraging code reuse. Batteries included :)
FuseMedML is part of the PyTorch Ecosystem.
- install instructions section
- complete code examples
- community support
- Contributing to FuseMedML guide
- citation info
Analyzing many ML research projects we discovered that
- Projects bring up is taking far too long, even when very similar projects were already done in the past by the same lab!
- Porting individual components across projects was painful - resulting in "reinventing the wheel" time after time
This is a key aspect in FuseMedML (shortly named as "fuse"). It's a key driver of flexibility, and allows to easily deal with multi modality information.
from fuse.utils import NDict
sample_ndict = NDict()
sample_ndict['input.mri'] = # ...
sample_ndict['input.ct_view_a'] = # ...
sample_ndict['input.ct_view_b'] = # ...
sample_ndict['groundtruth.disease_level_label'] = # ...This data can be a single sample, it can be for a minibatch, for an entire epoch, or anything that is desired. The "nested key" ("a.b.c.d.etc') is called "path key", as it can be seen as a path inside the nested dictionary.
Components are written in a way that allows to define input and output keys, to be read and written from the nested dict See a short introduction video (3 minutes) to how FuseMedML components work:
https://user-images.githubusercontent.com/7043815/177197158-d3ea0736-629e-4dcb-bd5e-666993fbcfa2.mp4
A multi head model FuseMedML style component, allows easy reuse across projects:
ModelMultiHead(
conv_inputs=(('data.input.img', 1),), # input to the backbone model
backbone=BackboneResnet3D(in_channels=1), # PyTorch nn Module
heads=[ # list of heads - gives the option to support multi task / multi head approach
Head3D(head_name='classification',
mode="classification",
conv_inputs=[("model.backbone_features", 512)] # Input to the classification head
,),
]
)Our default loss implementation - creates an easy wrap around a callable function, while being FuseMedML style
LossDefault(
pred='model.logits.classification', # input - model prediction scores
target='data.label', # input - ground truth labels
callable=torch.nn.functional.cross_entropy # callable - function that will get the prediction scores and labels extracted from batch_dict and compute the loss
)An example metric that can be used
MetricAUCROC(
pred='model.output', # input - model prediction scores
target='data.label' # input - ground truth labels
)Note that several components return answers directly and not write it into the nested dictionary. This is perfectly fine, and to allow maximum flexibility we do not require any usage of output path keys.
Creating custom FuseMedML components is easy - in the following example we add a new data processing operator:
A data pipeline operator
class OpPad(OpBase):
def __call__(self, sample_dict: NDict,
key_in: str,
padding: List[int], fill: int = 0, mode: str = 'constant',
key_out:Optional[str]=None,
):
# we extract the element in the defined key location (for example 'input.xray_img')
img = sample_dict[key_in]
assert isinstance(img, np.ndarray), f'Expected np.ndarray but got {type(img)}'
processed_img = np.pad(img, pad_width=padding, mode=mode, constant_values=fill)
# store the result in the requested output key (or in key_in if no key_out is provided)
key_out = key_in if key_out is None
sample_dict[key_out] = processed_img
# returned the modified nested dict
return sample_dictSince the key location isn't hardcoded, this module can be easily reused across different research projects with very different data sample structures. More code reuse - Hooray!
FuseMedML-style components in general are any classes or functions that define which key paths will be written and which will be read. Arguments can be freely named, and you don't even have to write anything to the nested dict. Some FuseMedML components return a value directly - for example, loss functions.
fuse.data - A declarative super flexible data processing pipeline
- Easy dealing with complex multi modality scenario
- Advanced caching, including periodic audits to automatically detect stale caches
- Default ready-to-use Dataset and Sampler classes
- See detailed introduction here
fuse.eval - a standalone library for evaluating ML models (not necessarily trained with FuseMedML)
The package includes collection of off-the-shelf metrics and utilities such as statistical significance tests, calibration, thresholding, model comparison and more. See detailed introduction here
fuse.dl - reusable dl (deep learning) model architecture components, loss functions, etc.
Some components depend on pytorch. For example, fuse.data is oriented towards pytorch DataSet, DataLoader, DataSampler etc.
fuse.dl makes heavy usage of pytorch models.
Some components do not depend on any specific DL library - for example fuse.eval.
Broadly speaking, the supported DL libraries are:
- "Pure" pytorch
- pytorch-lightning
Before you ask - pytorch-lightning and FuseMedML play along very nicely and have in practice orthogonal and additive benefits :) See Simple FuseMedML + PytorchLightning Example for simple supervised learning cases, and this example for completely custom usage of pytorch-lightning and FuseMedML - useful for advanced scenarios such as Reinforcement Learning and generative models.
fuse-med-ml, the core library, is completely domain agnostic! Domain extensions are optionally installable packages that deal with specific (sub) domains. For example:
- fuseimg which was battle-tested in many medical imaging related projects (different organs, imaging modalities, tasks, etc.)
- fusedrug (to be released soon) which focuses on molecular biology and chemistry - prediction, generation and more
Domain extensions contain concrete implementation of components and components parts within the relevant domain, for example:
- Data pipeline operations - for example, a 3d affine transformation of a 3d image
- Evaluation metrics - for example, a custom metric evaluating docking of a potential drug with a protein target
- Loss functions - for example, a custom segmentation evaluation loss
The recommended directory structure mimics fuse-med-ml core structure
your_package
data #everything related to datasets, samplers, data processing pipeline Ops, etc.
dl #everything related to deep learning architectures, optimizers, loss functions etc.
eval #evaluation metrics
utils #any utilities
You are highly encouraged to create additional domain extensions and/or contribute to the existing ones! There's no need to wait for any approval, you can create domain extensions on your own repos right away
Note - in general, we find it helpful to follow the same directory structure shown above even in small and specific research projects that use FuseMedML for consistency and easy landing for newcomers into your project :)
FuseMedML is tested on Python >= 3.9 and PyTorch >= 2.0
Create a conda environment using the following command (you can replace FUSEMEDML with your preferred enviornment name)
conda create -n FUSEMEDML python=3.9
conda activate FUSEMEDMLNow one shall install PyTorch and it's corresponding cudatoolkit. See here for the exact command that will suit your local environment. For example:
conda install pytorch torchvision torchaudio pytorch-cuda=11.6 -c pytorch -c nvidia
and then do Option 1 or Option 2 below inside the activated conda env
The best way to install FuseMedML is to clone the repository and install it in an editable mode using pip:
$ pip install -e .[all]This mode installs all the currently publicly available domain extensions - fuseimg as of now, fusedrug will be added soon.
To install FuseMedML with an included collection of examples install it using:
$ pip install -e .[all,examples]$ pip install fuse-med-ml[all]or with examples:
$ pip install fuse-med-ml[all,examples]- Easy access "Hello World" colab notebook
- Classification
-
MNIST - a simple example, including training, inference and evaluation over MNIST dataset
-
STOIC - severe COVID-19 classifier baseline given a Computed-Tomography (CT), age group and gender. Challenge description
-
KNIGHT Challenge - preoperative prediction of risk class for patients with renal masses identified in clinical Computed Tomography (CT) imaging of the kidneys. Including data pre-processing, baseline implementation and evaluation pipeline for the challenge.
-
Multimodality tutorial - demonstration of two popular simple methods integrating imaging and clinical data (tabular) using FuseMedML
-
Skin Lesion - skin lesion classification , including training, inference and evaluation over the public dataset introduced in ISIC challenge
-
Breast Cancer Lesion Classification - lesions classification of tumor ( benign, malignant) in breast mammography over the public dataset introduced in The Chinese Mammography Database (CMMD)
-
Mortality prediction for ICU patients - Example of EHR transformer applied to the data of Intensive Care Units patients for in-hospital mortality prediction. The dataset is from PhysioNet Computing in Cardiology Challenge (2012)
-
- Pre-training
- Medical Imaging Pre-training and Downstream Task Validation - pre-training a model on 3D MRI medical imaging and then using it for classification and segmentation downstream tasks.
- Walkthrough Template - includes several TODO notes, marking the minimal scope of code required to get your pipeline up and running. The template also includes useful explanations and tips.
- Slack workspace at fusemedml.slack.com for informal communication - click here to join
- Github Discussions
If you use FuseMedML in scientific context, please consider citing our JOSS paper:
@article{Golts2023,
doi = {10.21105/joss.04943},
url = {https://doi.org/10.21105/joss.04943},
year = {2023},
publisher = {The Open Journal},
volume = {8},
number = {81},
pages = {4943},
author = {Alex Golts and Moshe Raboh and Yoel Shoshan and Sagi Polaczek and Simona Rabinovici-Cohen and Efrat Hexter},
title = {FuseMedML: a framework for accelerated discovery in machine learning based biomedicine},
journal = {Journal of Open Source Software}
}For Tasks:
Click tags to check more tools for each tasksFor Jobs:
Alternative AI tools for fuse-med-ml
Similar Open Source Tools

fuse-med-ml
FuseMedML is a Python framework designed to accelerate machine learning-based discovery in the medical field by promoting code reuse. It provides a flexible design concept where data is stored in a nested dictionary, allowing easy handling of multi-modality information. The framework includes components for creating custom models, loss functions, metrics, and data processing operators. Additionally, FuseMedML offers 'batteries included' key components such as fuse.data for data processing, fuse.eval for model evaluation, and fuse.dl for reusable deep learning components. It supports PyTorch and PyTorch Lightning libraries and encourages the creation of domain extensions for specific medical domains.
codellm-devkit
Codellm-devkit (CLDK) is a Python library that serves as a multilingual program analysis framework bridging traditional static analysis tools and Large Language Models (LLMs) specialized for code (CodeLLMs). It simplifies the process of analyzing codebases across multiple programming languages, enabling the extraction of meaningful insights and facilitating LLM-based code analysis. The library provides a unified interface for integrating outputs from various analysis tools and preparing them for effective use by CodeLLMs. Codellm-devkit aims to enable the development and experimentation of robust analysis pipelines that combine traditional program analysis tools and CodeLLMs, reducing friction in multi-language code analysis and ensuring compatibility across different tools and LLM platforms. It is designed to seamlessly integrate with popular analysis tools like WALA, Tree-sitter, LLVM, and CodeQL, acting as a crucial intermediary layer for efficient communication between these tools and CodeLLMs. The project is continuously evolving to include new tools and frameworks, maintaining its versatility for code analysis and LLM integration.

Biomni
Biomni is a general-purpose biomedical AI agent designed to autonomously execute a wide range of research tasks across diverse biomedical subfields. By integrating cutting-edge large language model (LLM) reasoning with retrieval-augmented planning and code-based execution, Biomni helps scientists dramatically enhance research productivity and generate testable hypotheses.
premsql
PremSQL is an open-source library designed to help developers create secure, fully local Text-to-SQL solutions using small language models. It provides essential tools for building and deploying end-to-end Text-to-SQL pipelines with customizable components, ideal for secure, autonomous AI-powered data analysis. The library offers features like Local-First approach, Customizable Datasets, Robust Executors and Evaluators, Advanced Generators, Error Handling and Self-Correction, Fine-Tuning Support, and End-to-End Pipelines. Users can fine-tune models, generate SQL queries from natural language inputs, handle errors, and evaluate model performance against predefined metrics. PremSQL is extendible for customization and private data usage.

zshot
Zshot is a highly customizable framework for performing Zero and Few shot named entity and relationships recognition. It can be used for mentions extraction, wikification, zero and few shot named entity recognition, zero and few shot named relationship recognition, and visualization of zero-shot NER and RE extraction. The framework consists of two main components: the mentions extractor and the linker. There are multiple mentions extractors and linkers available, each serving a specific purpose. Zshot also includes a relations extractor and a knowledge extractor for extracting relations among entities and performing entity classification. The tool requires Python 3.6+ and dependencies like spacy, torch, transformers, evaluate, and datasets for evaluation over datasets like OntoNotes. Optional dependencies include flair and blink for additional functionalities. Zshot provides examples, tutorials, and evaluation methods to assess the performance of the components.
kafka-ml
Kafka-ML is a framework designed to manage the pipeline of Tensorflow/Keras and PyTorch machine learning models on Kubernetes. It enables the design, training, and inference of ML models with datasets fed through Apache Kafka, connecting them directly to data streams like those from IoT devices. The Web UI allows easy definition of ML models without external libraries, catering to both experts and non-experts in ML/AI.

embodied-agents
Embodied Agents is a toolkit for integrating large multi-modal models into existing robot stacks with just a few lines of code. It provides consistency, reliability, scalability, and is configurable to any observation and action space. The toolkit is designed to reduce complexities involved in setting up inference endpoints, converting between different model formats, and collecting/storing datasets. It aims to facilitate data collection and sharing among roboticists by providing Python-first abstractions that are modular, extensible, and applicable to a wide range of tasks. The toolkit supports asynchronous and remote thread-safe agent execution for maximal responsiveness and scalability, and is compatible with various APIs like HuggingFace Spaces, Datasets, Gymnasium Spaces, Ollama, and OpenAI. It also offers automatic dataset recording and optional uploads to the HuggingFace hub.
deep-research
Deep Research is a lightning-fast tool that uses powerful AI models to generate comprehensive research reports in just a few minutes. It leverages advanced 'Thinking' and 'Task' models, combined with an internet connection, to provide fast and insightful analysis on various topics. The tool ensures privacy by processing and storing all data locally. It supports multi-platform deployment, offers support for various large language models, web search functionality, knowledge graph generation, research history preservation, local and server API support, PWA technology, multi-key payload support, multi-language support, and is built with modern technologies like Next.js and Shadcn UI. Deep Research is open-source under the MIT License.
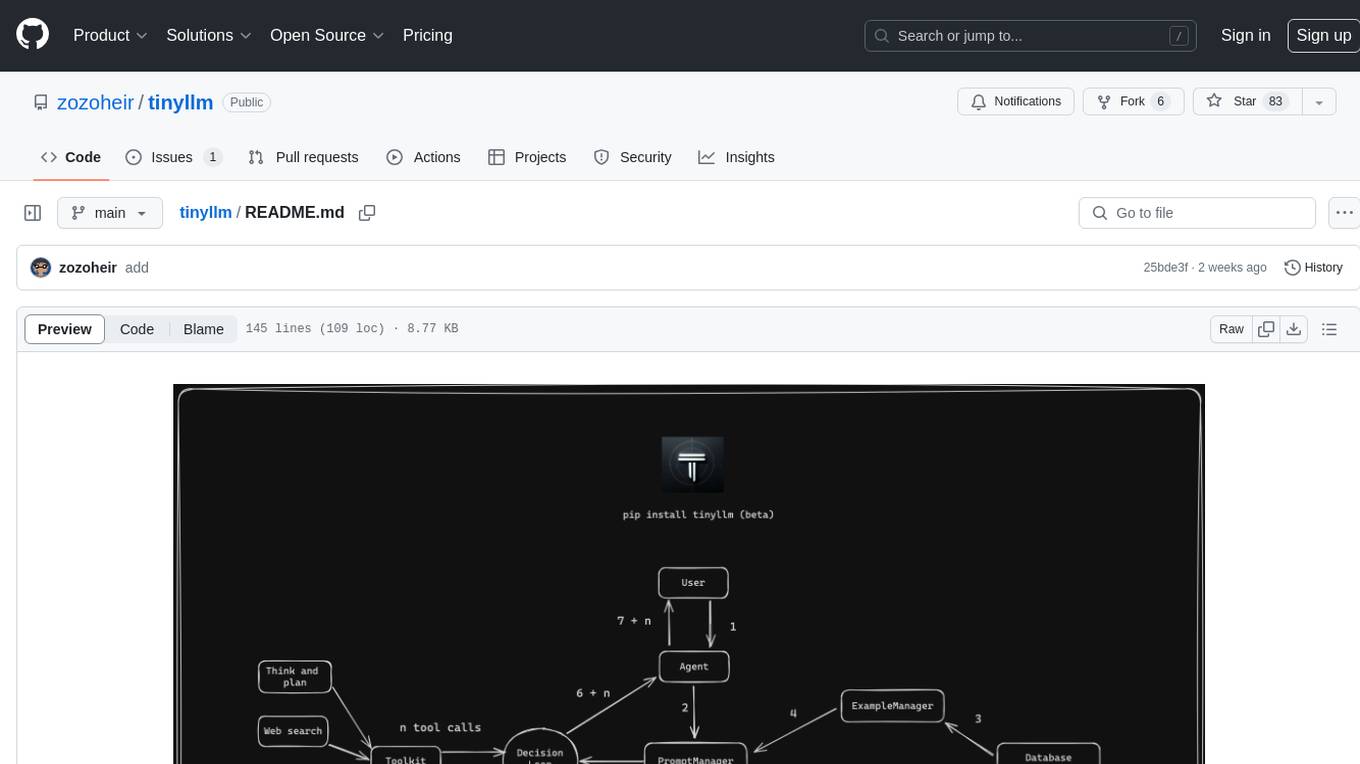
tinyllm
tinyllm is a lightweight framework designed for developing, debugging, and monitoring LLM and Agent powered applications at scale. It aims to simplify code while enabling users to create complex agents or LLM workflows in production. The core classes, Function and FunctionStream, standardize and control LLM, ToolStore, and relevant calls for scalable production use. It offers structured handling of function execution, including input/output validation, error handling, evaluation, and more, all while maintaining code readability. Users can create chains with prompts, LLM models, and evaluators in a single file without the need for extensive class definitions or spaghetti code. Additionally, tinyllm integrates with various libraries like Langfuse and provides tools for prompt engineering, observability, logging, and finite state machine design.
AI-Scientist
The AI Scientist is a comprehensive system for fully automatic scientific discovery, enabling Foundation Models to perform research independently. It aims to tackle the grand challenge of developing agents capable of conducting scientific research and discovering new knowledge. The tool generates papers on various topics using Large Language Models (LLMs) and provides a platform for exploring new research ideas. Users can create their own templates for specific areas of study and run experiments to generate papers. However, caution is advised as the codebase executes LLM-written code, which may pose risks such as the use of potentially dangerous packages and web access.
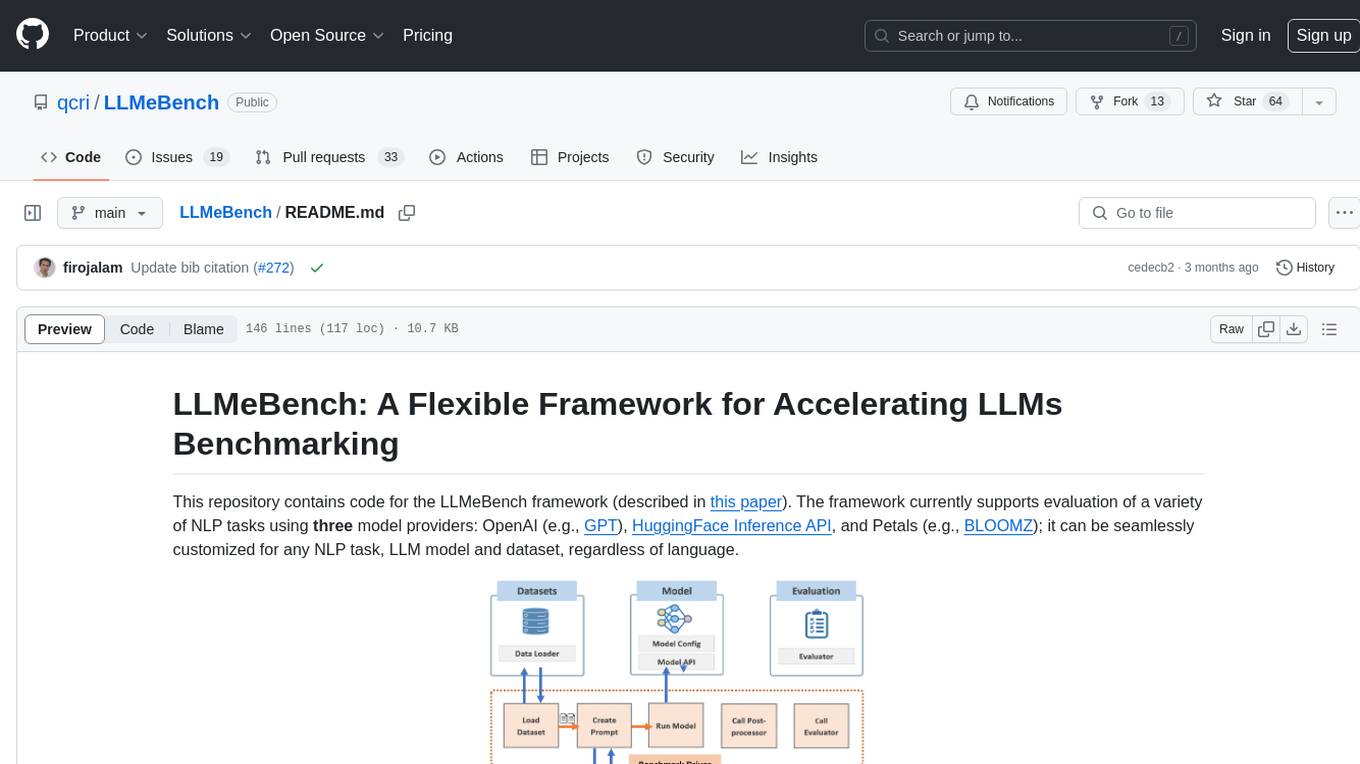
LLMeBench
LLMeBench is a flexible framework designed for accelerating benchmarking of Large Language Models (LLMs) in the field of Natural Language Processing (NLP). It supports evaluation of various NLP tasks using model providers like OpenAI, HuggingFace Inference API, and Petals. The framework is customizable for different NLP tasks, LLM models, and datasets across multiple languages. It features extensive caching capabilities, supports zero- and few-shot learning paradigms, and allows on-the-fly dataset download and caching. LLMeBench is open-source and continuously expanding to support new models accessible through APIs.
artkit
ARTKIT is a Python framework developed by BCG X for automating prompt-based testing and evaluation of Gen AI applications. It allows users to develop automated end-to-end testing and evaluation pipelines for Gen AI systems, supporting multi-turn conversations and various testing scenarios like Q&A accuracy, brand values, equitability, safety, and security. The framework provides a simple API, asynchronous processing, caching, model agnostic support, end-to-end pipelines, multi-turn conversations, robust data flows, and visualizations. ARTKIT is designed for customization by data scientists and engineers to enhance human-in-the-loop testing and evaluation, emphasizing the importance of tailored testing for each Gen AI use case.
neuron-ai
Neuron AI is a PHP framework that provides an Agent class for creating fully functional agents to perform tasks like analyzing text for SEO optimization. The framework manages advanced mechanisms such as memory, tools, and function calls. Users can extend the Agent class to create custom agents and interact with them to get responses based on the underlying LLM. Neuron AI aims to simplify the development of AI-powered applications by offering a structured framework with documentation and guidelines for contributions under the MIT license.
arbigent
Arbigent (Arbiter-Agent) is an AI agent testing framework designed to make AI agent testing practical for modern applications. It addresses challenges faced by traditional UI testing frameworks and AI agents by breaking down complex tasks into smaller, dependent scenarios. The framework is customizable for various AI providers, operating systems, and form factors, empowering users with extensive customization capabilities. Arbigent offers an intuitive UI for scenario creation and a powerful code interface for seamless test execution. It supports multiple form factors, optimizes UI for AI interaction, and is cost-effective by utilizing models like GPT-4o mini. With a flexible code interface and open-source nature, Arbigent aims to revolutionize AI agent testing in modern applications.
Trace
Trace is a new AutoDiff-like tool for training AI systems end-to-end with general feedback. It generalizes the back-propagation algorithm by capturing and propagating an AI system's execution trace. Implemented as a PyTorch-like Python library, users can write Python code directly and use Trace primitives to optimize certain parts, similar to training neural networks.
mosec
Mosec is a high-performance and flexible model serving framework for building ML model-enabled backend and microservices. It bridges the gap between any machine learning models you just trained and the efficient online service API. * **Highly performant** : web layer and task coordination built with Rust 🦀, which offers blazing speed in addition to efficient CPU utilization powered by async I/O * **Ease of use** : user interface purely in Python 🐍, by which users can serve their models in an ML framework-agnostic manner using the same code as they do for offline testing * **Dynamic batching** : aggregate requests from different users for batched inference and distribute results back * **Pipelined stages** : spawn multiple processes for pipelined stages to handle CPU/GPU/IO mixed workloads * **Cloud friendly** : designed to run in the cloud, with the model warmup, graceful shutdown, and Prometheus monitoring metrics, easily managed by Kubernetes or any container orchestration systems * **Do one thing well** : focus on the online serving part, users can pay attention to the model optimization and business logic
For similar tasks

fuse-med-ml
FuseMedML is a Python framework designed to accelerate machine learning-based discovery in the medical field by promoting code reuse. It provides a flexible design concept where data is stored in a nested dictionary, allowing easy handling of multi-modality information. The framework includes components for creating custom models, loss functions, metrics, and data processing operators. Additionally, FuseMedML offers 'batteries included' key components such as fuse.data for data processing, fuse.eval for model evaluation, and fuse.dl for reusable deep learning components. It supports PyTorch and PyTorch Lightning libraries and encourages the creation of domain extensions for specific medical domains.
For similar jobs
grand-challenge.org
Grand Challenge is a platform that provides access to large amounts of annotated training data, objective comparisons of state-of-the-art machine learning solutions, and clinical validation using real-world data. It assists researchers, data scientists, and clinicians in collaborating to develop robust machine learning solutions to problems in biomedical imaging.
Detection-and-Classification-of-Alzheimers-Disease
This tool is designed to detect and classify Alzheimer's Disease using Deep Learning and Machine Learning algorithms on an early basis, which is further optimized using the Crow Search Algorithm (CSA). Alzheimer's is a fatal disease, and early detection is crucial for patients to predetermine their condition and prevent its progression. By analyzing MRI scanned images using Artificial Intelligence technology, this tool can classify patients who may or may not develop AD in the future. The CSA algorithm, combined with ML algorithms, has proven to be the most effective approach for this purpose.

OpenCRISPR
OpenCRISPR is a set of free and open gene editing systems designed by Profluent Bio. The OpenCRISPR-1 protein maintains the prototypical architecture of a Type II Cas9 nuclease but is hundreds of mutations away from SpCas9 or any other known natural CRISPR-associated protein. You can view OpenCRISPR-1 as a drop-in replacement for many protocols that need a cas9-like protein with an NGG PAM and you can even use it with canonical SpCas9 gRNAs. OpenCRISPR-1 can be fused in a deactivated or nickase format for next generation gene editing techniques like base, prime, or epigenome editing.

AlphaFold3
AlphaFold3 is an implementation of the Alpha Fold 3 model in PyTorch for accurate structure prediction of biomolecular interactions. It includes modules for genetic diffusion and full model examples for forward pass computations. The tool allows users to generate random pair and single representations, operate on atomic coordinates, and perform structure predictions based on input tensors. The implementation also provides functionalities for training and evaluating the model.

fuse-med-ml
FuseMedML is a Python framework designed to accelerate machine learning-based discovery in the medical field by promoting code reuse. It provides a flexible design concept where data is stored in a nested dictionary, allowing easy handling of multi-modality information. The framework includes components for creating custom models, loss functions, metrics, and data processing operators. Additionally, FuseMedML offers 'batteries included' key components such as fuse.data for data processing, fuse.eval for model evaluation, and fuse.dl for reusable deep learning components. It supports PyTorch and PyTorch Lightning libraries and encourages the creation of domain extensions for specific medical domains.
hi-ml
The Microsoft Health Intelligence Machine Learning Toolbox is a repository that provides low-level and high-level building blocks for Machine Learning / AI researchers and practitioners. It simplifies and streamlines work on deep learning models for healthcare and life sciences by offering tested components such as data loaders, pre-processing tools, deep learning models, and cloud integration utilities. The repository includes two Python packages, 'hi-ml-azure' for helper functions in AzureML, 'hi-ml' for ML components, and 'hi-ml-cpath' for models and workflows related to histopathology images.
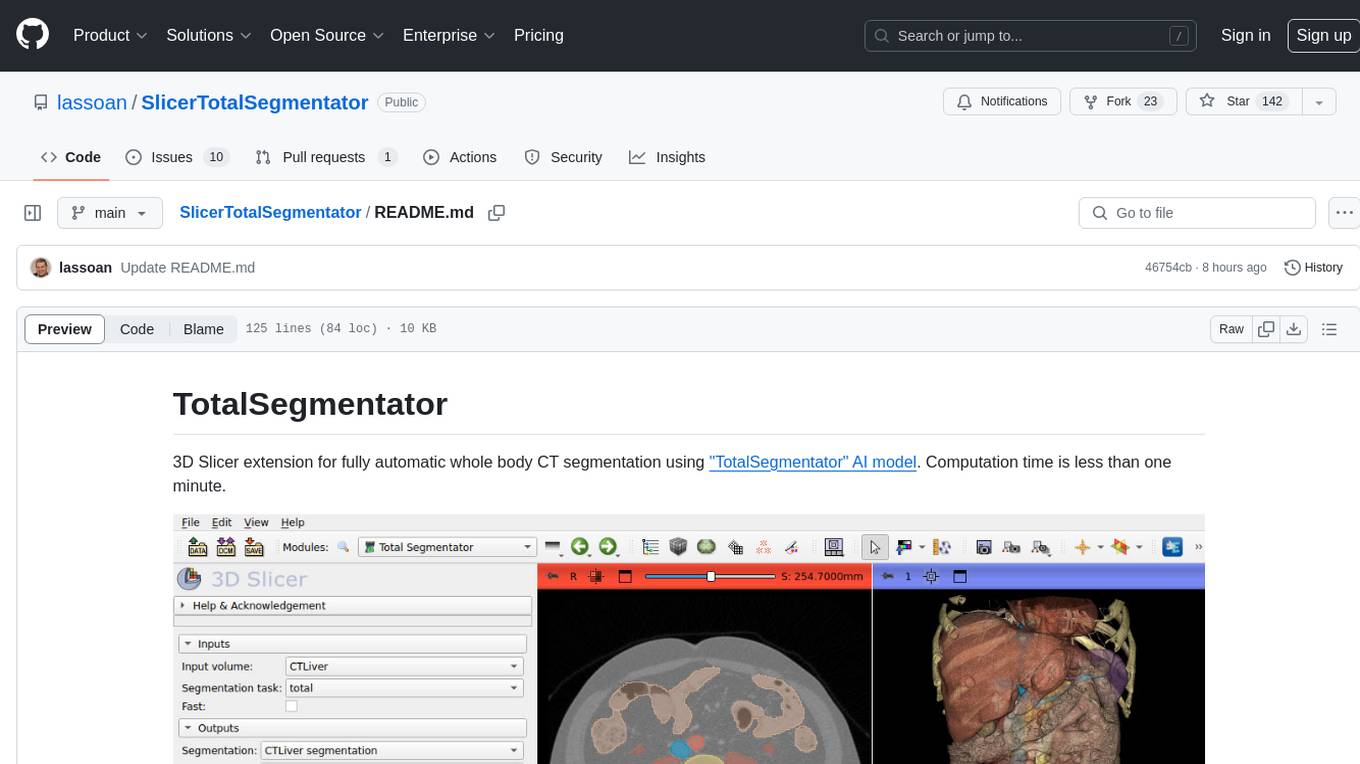
SlicerTotalSegmentator
TotalSegmentator is a 3D Slicer extension designed for fully automatic whole body CT segmentation using the 'TotalSegmentator' AI model. The computation time is less than one minute, making it efficient for research purposes. Users can set up GPU acceleration for faster segmentation. The tool provides a user-friendly interface for loading CT images, creating segmentations, and displaying results in 3D. Troubleshooting steps are available for common issues such as failed computation, GPU errors, and inaccurate segmentations. Contributions to the extension are welcome, following 3D Slicer contribution guidelines.
md-agent
MD-Agent is a LLM-agent based toolset for Molecular Dynamics. It uses Langchain and a collection of tools to set up and execute molecular dynamics simulations, particularly in OpenMM. The tool assists in environment setup, installation, and usage by providing detailed steps. It also requires API keys for certain functionalities, such as OpenAI and paper-qa for literature searches. Contributions to the project are welcome, with a detailed Contributor's Guide available for interested individuals.


