
crystal-text-llm
Large language models to generate stable crystals.
Stars: 54
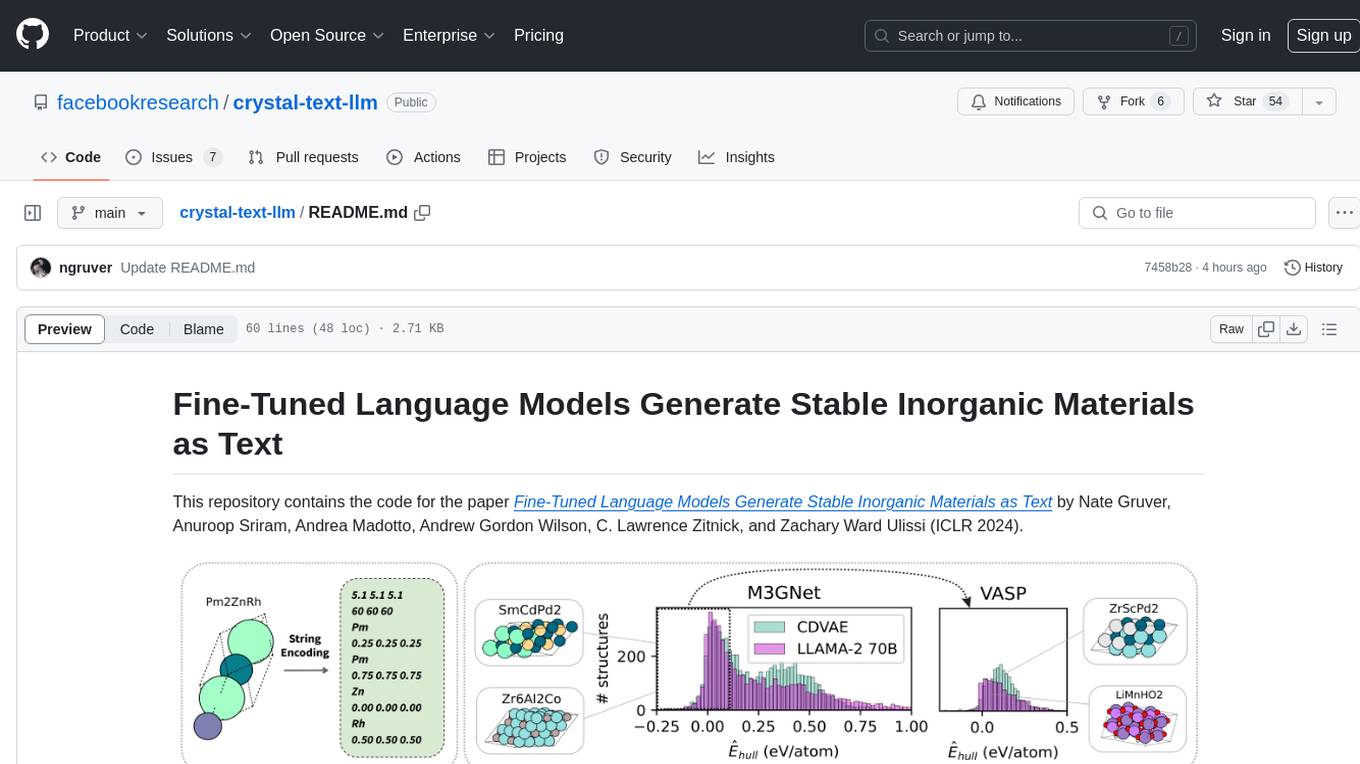
This repository contains the code for the paper Fine-Tuned Language Models Generate Stable Inorganic Materials as Text. It demonstrates how finetuned LLMs can be used to generate stable materials, match or exceed the performance of domain specific models, mutate existing materials, and sample crystal structures conditioned on text descriptions. The method is distinct from CrystaLLM, which trains language models from scratch on CIF-formatted crystals.
README:
This repository contains the code for the paper Fine-Tuned Language Models Generate Stable Inorganic Materials as Text by Nate Gruver, Anuroop Sriram, Andrea Madotto, Andrew Gordon Wilson, C. Lawrence Zitnick, and Zachary Ward Ulissi (ICLR 2024).
We show that finetuned LLMs can be used to generate stable materials using string encodings. These finetuned LLMs can match or exceed the performance of a domain specific diffusion model (CDVAE). LLMs can also be used to mutate existing materials or to sample crystal structures conditioned on text descriptions.
Run the following command to install all dependencies.
source install.sh
After installation, activate the environment with
conda activate crystal-llm
If you prefer not using conda, you can also install the dependencies listed in install.sh manually.
Run training with
python llama_finetune.py --run-name 7b-test-run --model 7b
and sample from a PEFT model with
python llama_sample.py --model_name 7b --model_path=exp/7b-test-run/checkpoint-500 --out_path=llm_samples.csv
The majority of crystall-llm is licensed under CC-BY-NC, however portions of the project are available under separate license terms: https://github.com/materialsproject/pymatgen is licensed under the MIT license, https://github.com/huggingface/transformers is licensed under Apache 2.0, and https://gitlab.com/ase/ase/-/ is licensed under GNU Lesser General License
Please cite our work as:
@inproceedings{gruver2023llmtime,
title={Fine-Tuned Language Models Generate Stable Inorganic Materials as Text},
author={Nate Gruver, Anuroop Sriram, Andrea Madotto, Andrew Gordon Wilson, C. Lawrence Zitnick, and Zachary Ward Ulissi},
booktitle={International Conference on Learning Representations 2024},
year={2024}
}For Tasks:
Click tags to check more tools for each tasksFor Jobs:
Alternative AI tools for crystal-text-llm
Similar Open Source Tools

crystal-text-llm
This repository contains the code for the paper Fine-Tuned Language Models Generate Stable Inorganic Materials as Text. It demonstrates how finetuned LLMs can be used to generate stable materials, match or exceed the performance of domain specific models, mutate existing materials, and sample crystal structures conditioned on text descriptions. The method is distinct from CrystaLLM, which trains language models from scratch on CIF-formatted crystals.
ModernBERT
ModernBERT is a repository focused on modernizing BERT through architecture changes and scaling. It introduces FlexBERT, a modular approach to encoder building blocks, and heavily relies on .yaml configuration files to build models. The codebase builds upon MosaicBERT and incorporates Flash Attention 2. The repository is used for pre-training and GLUE evaluations, with a focus on reproducibility and documentation. It provides a collaboration between Answer.AI, LightOn, and friends.
xlstm-jax
The xLSTM-jax repository contains code for training and evaluating the xLSTM model on language modeling using JAX. xLSTM is a Recurrent Neural Network architecture that improves upon the original LSTM through Exponential Gating, normalization, stabilization techniques, and a Matrix Memory. It is optimized for large-scale distributed systems with performant triton kernels for faster training and inference.
LangSim
LangSim is a tool developed to address the challenge of using simulation tools in computational chemistry and materials science, which typically require cryptic input files or programming experience. The tool provides a Large Language Model (LLM) extension with agents to couple the LLM to scientific simulation codes and calculate physical properties from a natural language interface. It aims to simplify the process of interacting with simulation tools by enabling users to query the large language model directly from a Python environment or a web-based interface.
aligner
Aligner is a model-agnostic alignment tool designed to efficiently correct responses from large language models. It redistributes initial answers to align with human intentions, improving performance across various LLMs. The tool can be applied with minimal training, enhancing upstream models and reducing hallucination. Aligner's 'copy and correct' method preserves the base structure while enhancing responses. It achieves significant performance improvements in helpfulness, harmlessness, and honesty dimensions, with notable success in boosting Win Rates on evaluation leaderboards.
LLMs-World-Models-for-Planning
This repository provides a Python implementation of a method that leverages pre-trained large language models to construct and utilize world models for model-based task planning. It includes scripts to generate domain models using natural language descriptions, correct domain models based on feedback, and support plan generation for tasks in different domains. The code has been refactored for better readability and includes tools for validating PDDL syntax and handling corrective feedback.
md-agent
MD-Agent is a LLM-agent based toolset for Molecular Dynamics. It uses Langchain and a collection of tools to set up and execute molecular dynamics simulations, particularly in OpenMM. The tool assists in environment setup, installation, and usage by providing detailed steps. It also requires API keys for certain functionalities, such as OpenAI and paper-qa for literature searches. Contributions to the project are welcome, with a detailed Contributor's Guide available for interested individuals.

Me-LLaMA
Me LLaMA introduces a suite of open-source medical Large Language Models (LLMs), including Me LLaMA 13B/70B and their chat-enhanced versions. Developed through innovative continual pre-training and instruction tuning, these models leverage a vast medical corpus comprising PubMed papers, medical guidelines, and general domain data. Me LLaMA sets new benchmarks on medical reasoning tasks, making it a significant asset for medical NLP applications and research. The models are intended for computational linguistics and medical research, not for clinical decision-making without validation and regulatory approval.
dbrx
DBRX is a large language model trained by Databricks and made available under an open license. It is a Mixture-of-Experts (MoE) model with 132B total parameters and 36B live parameters, using 16 experts, of which 4 are active during training or inference. DBRX was pre-trained for 12T tokens of text and has a context length of 32K tokens. The model is available in two versions: a base model and an Instruct model, which is finetuned for instruction following. DBRX can be used for a variety of tasks, including text generation, question answering, summarization, and translation.

RAG-FiT
RAG-FiT is a library designed to improve Language Models' ability to use external information by fine-tuning models on specially created RAG-augmented datasets. The library assists in creating training data, training models using parameter-efficient finetuning (PEFT), and evaluating performance using RAG-specific metrics. It is modular, customizable via configuration files, and facilitates fast prototyping and experimentation with various RAG settings and configurations.
spear
SPEAR (Simulator for Photorealistic Embodied AI Research) is a powerful tool for training embodied agents. It features 300 unique virtual indoor environments with 2,566 unique rooms and 17,234 unique objects that can be manipulated individually. Each environment is designed by a professional artist and features detailed geometry, photorealistic materials, and a unique floor plan and object layout. SPEAR is implemented as Unreal Engine assets and provides an OpenAI Gym interface for interacting with the environments via Python.
hi-ml
The Microsoft Health Intelligence Machine Learning Toolbox is a repository that provides low-level and high-level building blocks for Machine Learning / AI researchers and practitioners. It simplifies and streamlines work on deep learning models for healthcare and life sciences by offering tested components such as data loaders, pre-processing tools, deep learning models, and cloud integration utilities. The repository includes two Python packages, 'hi-ml-azure' for helper functions in AzureML, 'hi-ml' for ML components, and 'hi-ml-cpath' for models and workflows related to histopathology images.
awesome-synthetic-datasets
This repository focuses on organizing resources for building synthetic datasets using large language models. It covers important datasets, libraries, tools, tutorials, and papers related to synthetic data generation. The goal is to provide pragmatic and practical resources for individuals interested in creating synthetic datasets for machine learning applications.

aitlas
The AiTLAS toolbox (Artificial Intelligence Toolbox for Earth Observation) includes state-of-the-art machine learning methods for exploratory and predictive analysis of satellite imagery as well as a repository of AI-ready Earth Observation (EO) datasets. It can be easily applied for a variety of Earth Observation tasks, such as land use and cover classification, crop type prediction, localization of specific objects (semantic segmentation), etc. The main goal of AiTLAS is to facilitate better usability and adoption of novel AI methods (and models) by EO experts, while offering easy access and standardized format of EO datasets to AI experts which allows benchmarking of various existing and novel AI methods tailored for EO data.

RAGFoundry
RAG Foundry is a library designed to enhance Large Language Models (LLMs) by fine-tuning models on RAG-augmented datasets. It helps create training data, train models using parameter-efficient finetuning (PEFT), and measure performance using RAG-specific metrics. The library is modular, customizable using configuration files, and facilitates prototyping with various RAG settings and configurations for tasks like data processing, retrieval, training, inference, and evaluation.

Chinese-Tiny-LLM
Chinese-Tiny-LLM is a repository containing procedures for cleaning Chinese web corpora and pre-training code. It introduces CT-LLM, a 2B parameter language model focused on the Chinese language. The model primarily uses Chinese data from a 1,200 billion token corpus, showing excellent performance in Chinese language tasks. The repository includes tools for filtering, deduplication, and pre-training, aiming to encourage further research and innovation in language model development.
For similar tasks

crystal-text-llm
This repository contains the code for the paper Fine-Tuned Language Models Generate Stable Inorganic Materials as Text. It demonstrates how finetuned LLMs can be used to generate stable materials, match or exceed the performance of domain specific models, mutate existing materials, and sample crystal structures conditioned on text descriptions. The method is distinct from CrystaLLM, which trains language models from scratch on CIF-formatted crystals.
llm-past-tense
The 'llm-past-tense' repository contains code related to the research paper 'Does Refusal Training in LLMs Generalize to the Past Tense?' by Maksym Andriushchenko and Nicolas Flammarion. It explores the generalization of refusal training in large language models (LLMs) to the past tense. The code includes experiments and examples for running different models and requests related to the study. Users can cite the work if found useful in their research, and the codebase is released under the MIT License.
ai-on-gke
This repository contains assets related to AI/ML workloads on Google Kubernetes Engine (GKE). Run optimized AI/ML workloads with Google Kubernetes Engine (GKE) platform orchestration capabilities. A robust AI/ML platform considers the following layers: Infrastructure orchestration that support GPUs and TPUs for training and serving workloads at scale Flexible integration with distributed computing and data processing frameworks Support for multiple teams on the same infrastructure to maximize utilization of resources

ray
Ray is a unified framework for scaling AI and Python applications. It consists of a core distributed runtime and a set of AI libraries for simplifying ML compute, including Data, Train, Tune, RLlib, and Serve. Ray runs on any machine, cluster, cloud provider, and Kubernetes, and features a growing ecosystem of community integrations. With Ray, you can seamlessly scale the same code from a laptop to a cluster, making it easy to meet the compute-intensive demands of modern ML workloads.

labelbox-python
Labelbox is a data-centric AI platform for enterprises to develop, optimize, and use AI to solve problems and power new products and services. Enterprises use Labelbox to curate data, generate high-quality human feedback data for computer vision and LLMs, evaluate model performance, and automate tasks by combining AI and human-centric workflows. The academic & research community uses Labelbox for cutting-edge AI research.
djl
Deep Java Library (DJL) is an open-source, high-level, engine-agnostic Java framework for deep learning. It is designed to be easy to get started with and simple to use for Java developers. DJL provides a native Java development experience and allows users to integrate machine learning and deep learning models with their Java applications. The framework is deep learning engine agnostic, enabling users to switch engines at any point for optimal performance. DJL's ergonomic API interface guides users with best practices to accomplish deep learning tasks, such as running inference and training neural networks.

mojo
Mojo is a new programming language that bridges the gap between research and production by combining Python syntax and ecosystem with systems programming and metaprogramming features. Mojo is still young, but it is designed to become a superset of Python over time.
tt-metal
TT-NN is a python & C++ Neural Network OP library. It provides a low-level programming model, TT-Metalium, enabling kernel development for Tenstorrent hardware.
For similar jobs
matsciml
The Open MatSci ML Toolkit is a flexible framework for machine learning in materials science. It provides a unified interface to a variety of materials science datasets, as well as a set of tools for data preprocessing, model training, and evaluation. The toolkit is designed to be easy to use for both beginners and experienced researchers, and it can be used to train models for a wide range of tasks, including property prediction, materials discovery, and materials design.

NoLabs
NoLabs is an open-source biolab that provides easy access to state-of-the-art models for bio research. It supports various tasks, including drug discovery, protein analysis, and small molecule design. NoLabs aims to accelerate bio research by making inference models accessible to everyone.

AlphaFold3
AlphaFold3 is an implementation of the Alpha Fold 3 model in PyTorch for accurate structure prediction of biomolecular interactions. It includes modules for genetic diffusion and full model examples for forward pass computations. The tool allows users to generate random pair and single representations, operate on atomic coordinates, and perform structure predictions based on input tensors. The implementation also provides functionalities for training and evaluating the model.

crystal-text-llm
This repository contains the code for the paper Fine-Tuned Language Models Generate Stable Inorganic Materials as Text. It demonstrates how finetuned LLMs can be used to generate stable materials, match or exceed the performance of domain specific models, mutate existing materials, and sample crystal structures conditioned on text descriptions. The method is distinct from CrystaLLM, which trains language models from scratch on CIF-formatted crystals.

Scientific-LLM-Survey
Scientific Large Language Models (Sci-LLMs) is a repository that collects papers on scientific large language models, focusing on biology and chemistry domains. It includes textual, molecular, protein, and genomic languages, as well as multimodal language. The repository covers various large language models for tasks such as molecule property prediction, interaction prediction, protein sequence representation, protein sequence generation/design, DNA-protein interaction prediction, and RNA prediction. It also provides datasets and benchmarks for evaluating these models. The repository aims to facilitate research and development in the field of scientific language modeling.

md-agent
MD-Agent is a LLM-agent based toolset for Molecular Dynamics. It uses Langchain and a collection of tools to set up and execute molecular dynamics simulations, particularly in OpenMM. The tool assists in environment setup, installation, and usage by providing detailed steps. It also requires API keys for certain functionalities, such as OpenAI and paper-qa for literature searches. Contributions to the project are welcome, with a detailed Contributor's Guide available for interested individuals.
AIMNet2
AIMNet2 Calculator is a package that integrates the AIMNet2 neural network potential into simulation workflows, providing fast and reliable energy, force, and property calculations for molecules with diverse elements. It excels at modeling various systems, offers flexible interfaces for popular simulation packages, and supports long-range interactions using DSF or Ewald summation Coulomb models. The tool is designed for accurate and versatile molecular simulations, suitable for large molecules and periodic calculations.
admet_ai
ADMET-AI is a platform for ADMET prediction using Chemprop-RDKit models trained on ADMET datasets from the Therapeutics Data Commons. It offers command line, Python API, and web server interfaces for making ADMET predictions on new molecules. The platform can be easily installed using pip and supports GPU acceleration. It also provides options for processing TDC data, plotting results, and hosting a web server. ADMET-AI is a machine learning platform for evaluating large-scale chemical libraries.
