
ontogpt
LLM-based ontological extraction tools, including SPIRES
Stars: 584
OntoGPT is a Python package for extracting structured information from text using large language models, instruction prompts, and ontology-based grounding. It provides a command line interface and a minimal web app for easy usage. The tool has been evaluated on test data and is used in related projects like TALISMAN for gene set analysis. OntoGPT enables users to extract information from text by specifying relevant terms and provides the extracted objects as output.
README:
OntoGPT is a Python package for extracting structured information from text with large language models (LLMs), instruction prompts, and ontology-based grounding.
For more details, please see the full documentation.
OntoGPT runs on the command line, though there's also a minimal web app interface (see Web Application section below).
-
Ensure you have Python 3.9 or greater installed.
-
Install with
pip:pip install ontogpt
-
Set your OpenAI API key:
runoak set-apikey -e openai <your openai api key>
-
See the list of all OntoGPT commands:
ontogpt --help
-
Try a simple example of information extraction:
echo "One treatment for high blood pressure is carvedilol." > example.txt ontogpt extract -i example.txt -t drug
OntoGPT will retrieve the necessary ontologies and output results to the command line. Your output will provide all extracted objects under the heading
extracted_object.
There is a bare bones web application for running OntoGPT and viewing results.
First, install the required dependencies with pip by running the following command:
pip install ontogpt[web]Then run this command to start the web application:
web-ontogptNOTE: We do not recommend hosting this webapp publicly without authentication.
OntoGPT uses the litellm package (https://litellm.vercel.app/) to interface with LLMs.
This means most APIs are supported, including OpenAI, Azure, Anthropic, Mistral, Replicate, and beyond.
The model name to use may be found from the command ontogpt list-models - use the name in the first column with the --model option.
In most cases, this will require setting the API key for a particular service as above:
runoak set-apikey -e anthropic-key <your anthropic api key>Some endpoints, such as OpenAI models through Azure, require setting additional details. These may be set similarly:
runoak set-apikey -e azure-key <your azure api key>
runoak set-apikey -e azure-base <your azure endpoint url>
runoak set-apikey -e azure-version <your azure api version, e.g. "2023-05-15">These details may also be set as environment variables as follows:
export AZURE_API_KEY="my-azure-api-key"
export AZURE_API_BASE="https://example-endpoint.openai.azure.com"
export AZURE_API_VERSION="2023-05-15"Open LLMs may be retrieved and run through the ollama package (https://ollama.com/).
You will need to install ollama (see the GitHub repo), and you may need to start it as a service with a command like ollama serve or sudo systemctl start ollama.
Then retrieve a model with ollama pull <modelname>, e.g., ollama pull llama3.
The model may then be used in OntoGPT by prefixing its name with ollama/, e.g., ollama/llama3, along with the --model option.
Some ollama models may not be listed in ontogpt list-models but the full list of downloaded LLMs can be seen with ollama list command.
OntoGPT's functions have been evaluated on test data. Please see the full documentation for details on these evaluations and how to reproduce them.
- TALISMAN, a tool for generating summaries of functions enriched within a gene set. TALISMAN uses OntoGPT to work with LLMs.
- Presentation: "Staying grounded: assembling structured biological knowledge with help from large language models" - presented by Harry Caufield as part of the AgBioData Consortium webinar series (September 2023)
- Presentation: "Transforming unstructured biomedical texts with large language models" - presented by Harry Caufield as part of the BOSC track at ISMB/ECCB 2023 (July 2023)
- Presentation: "OntoGPT: A framework for working with ontologies and large language models" - talk by Chris Mungall at Joint Food Ontology Workgroup (May 2023)
The information extraction approach used in OntoGPT, SPIRES, is described further in: Caufield JH, Hegde H, Emonet V, Harris NL, Joachimiak MP, Matentzoglu N, et al. Structured prompt interrogation and recursive extraction of semantics (SPIRES): A method for populating knowledge bases using zero-shot learning. Bioinformatics, Volume 40, Issue 3, March 2024, btae104, https://doi.org/10.1093/bioinformatics/btae104.
This project is part of the Monarch Initiative. We also gratefully acknowledge Bosch Research for their support of this research project.
For Tasks:
Click tags to check more tools for each tasksFor Jobs:
Alternative AI tools for ontogpt
Similar Open Source Tools

ontogpt
OntoGPT is a Python package for extracting structured information from text using large language models, instruction prompts, and ontology-based grounding. It provides a command line interface and a minimal web app for easy usage. The tool has been evaluated on test data and is used in related projects like TALISMAN for gene set analysis. OntoGPT enables users to extract information from text by specifying relevant terms and provides the extracted objects as output.
2p-kt
2P-Kt is a Kotlin-based and multi-platform reboot of tuProlog (2P), a multi-paradigm logic programming framework written in Java. It consists of an open ecosystem for Symbolic Artificial Intelligence (AI) with modules supporting logic terms, unification, indexing, resolution of logic queries, probabilistic logic programming, binary decision diagrams, OR-concurrent resolution, DSL for logic programming, parsing modules, serialisation modules, command-line interface, and graphical user interface. The tool is designed to support knowledge representation and automatic reasoning through logic programming in an extensible and flexible way, encouraging extensions towards other symbolic AI systems than Prolog. It is a pure, multi-platform Kotlin project supporting JVM, JS, Android, and Native platforms, with a lightweight library leveraging the Kotlin common library.
BTGenBot
BTGenBot is a tool that generates behavior trees for robots using lightweight large language models (LLMs) with a maximum of 7 billion parameters. It fine-tunes on a specific dataset, compares multiple LLMs, and evaluates generated behavior trees using various methods. The tool demonstrates the potential of LLMs with a limited number of parameters in creating effective and efficient robot behaviors.

MegatronApp
MegatronApp is a toolchain built around the Megatron-LM training framework, offering performance tuning, slow-node detection, and training-process visualization. It includes modules like MegaScan for anomaly detection, MegaFBD for forward-backward decoupling, MegaDPP for dynamic pipeline planning, and MegaScope for visualization. The tool aims to enhance large-scale distributed training by providing valuable capabilities and insights.
POPPER
Popper is an agentic framework for automated validation of free-form hypotheses using Large Language Models (LLMs). It follows Karl Popper's principle of falsification and designs falsification experiments to validate hypotheses. Popper ensures strict Type-I error control and actively gathers evidence from diverse observations. It delivers robust error control, high power, and scalability across various domains like biology, economics, and sociology. Compared to human scientists, Popper achieves comparable performance in validating complex biological hypotheses while reducing time by 10 folds, providing a scalable, rigorous solution for hypothesis validation.
curate-gpt
CurateGPT is a prototype web application and framework for performing general purpose AI-guided curation and curation-related operations over collections of objects. It allows users to load JSON, YAML, or CSV data, build vector database indexes for ontologies, and interact with various data sources like GitHub, Google Drives, Google Sheets, and more. The tool supports ontology curation, knowledge base querying, term autocompletion, and all-by-all comparisons for objects in a collection.
LLM-Merging
LLM-Merging is a repository containing starter code for the LLM-Merging competition. It provides a platform for efficiently building LLMs through merging methods. Users can develop new merging methods by creating new files in the specified directory and extending existing classes. The repository includes instructions for setting up the environment, developing new merging methods, testing the methods on specific datasets, and submitting solutions for evaluation. It aims to facilitate the development and evaluation of merging methods for LLMs.
curategpt
CurateGPT is a prototype web application and framework designed for general purpose AI-guided curation and curation-related operations over collections of objects. It provides functionalities for loading example data, building indexes, interacting with knowledge bases, and performing tasks such as chatting with a knowledge base, querying Pubmed, interacting with a GitHub issue tracker, term autocompletion, and all-by-all comparisons. The tool is built to work best with the OpenAI gpt-4 model and OpenAI ada-text-embedding-002 for embedding, but also supports alternative models through a plugin architecture.
monitors4codegen
This repository hosts the official code and data artifact for the paper 'Monitor-Guided Decoding of Code LMs with Static Analysis of Repository Context'. It introduces Monitor-Guided Decoding (MGD) for code generation using Language Models, where a monitor uses static analysis to guide the decoding. The repository contains datasets, evaluation scripts, inference results, a language server client 'multilspy' for static analyses, and implementation of various monitors monitoring for different properties in 3 programming languages. The monitors guide Language Models to adhere to properties like valid identifier dereferences, correct number of arguments to method calls, typestate validity of method call sequences, and more.
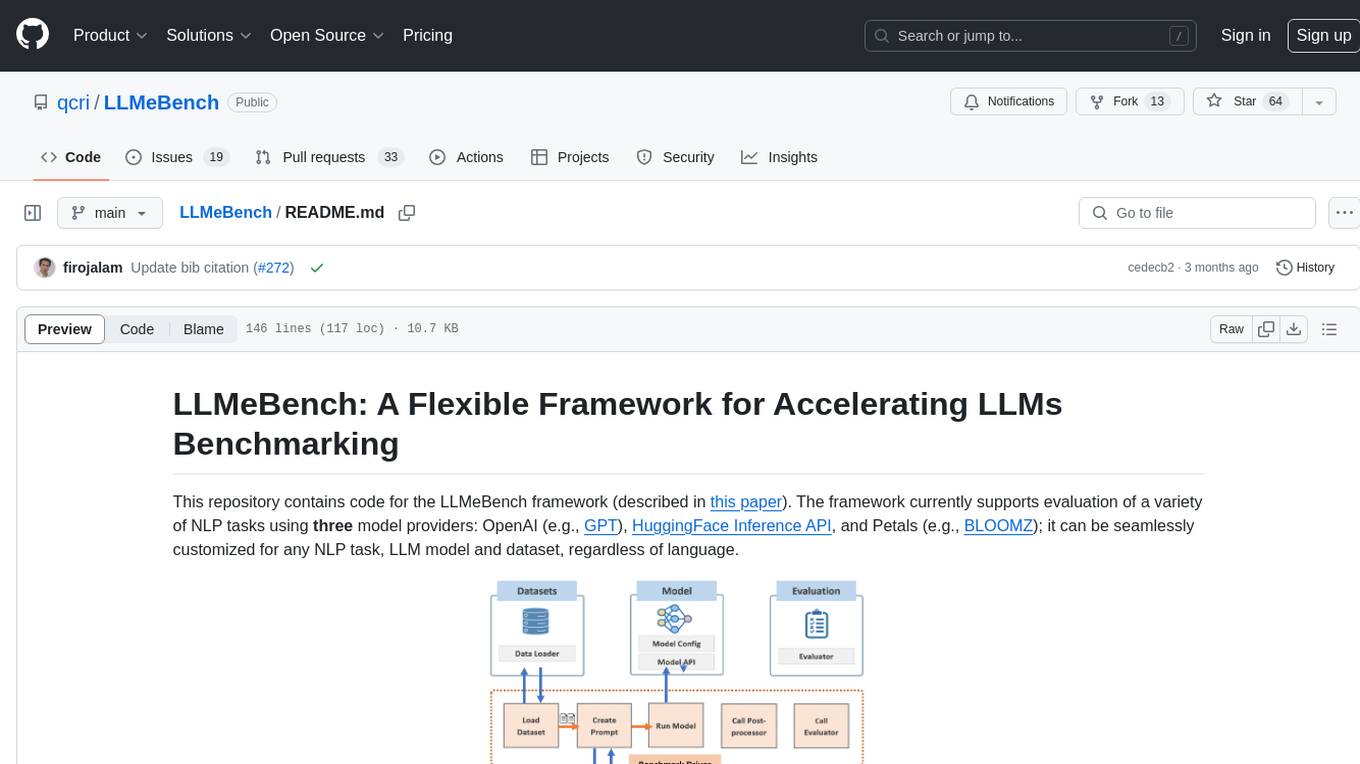
LLMeBench
LLMeBench is a flexible framework designed for accelerating benchmarking of Large Language Models (LLMs) in the field of Natural Language Processing (NLP). It supports evaluation of various NLP tasks using model providers like OpenAI, HuggingFace Inference API, and Petals. The framework is customizable for different NLP tasks, LLM models, and datasets across multiple languages. It features extensive caching capabilities, supports zero- and few-shot learning paradigms, and allows on-the-fly dataset download and caching. LLMeBench is open-source and continuously expanding to support new models accessible through APIs.
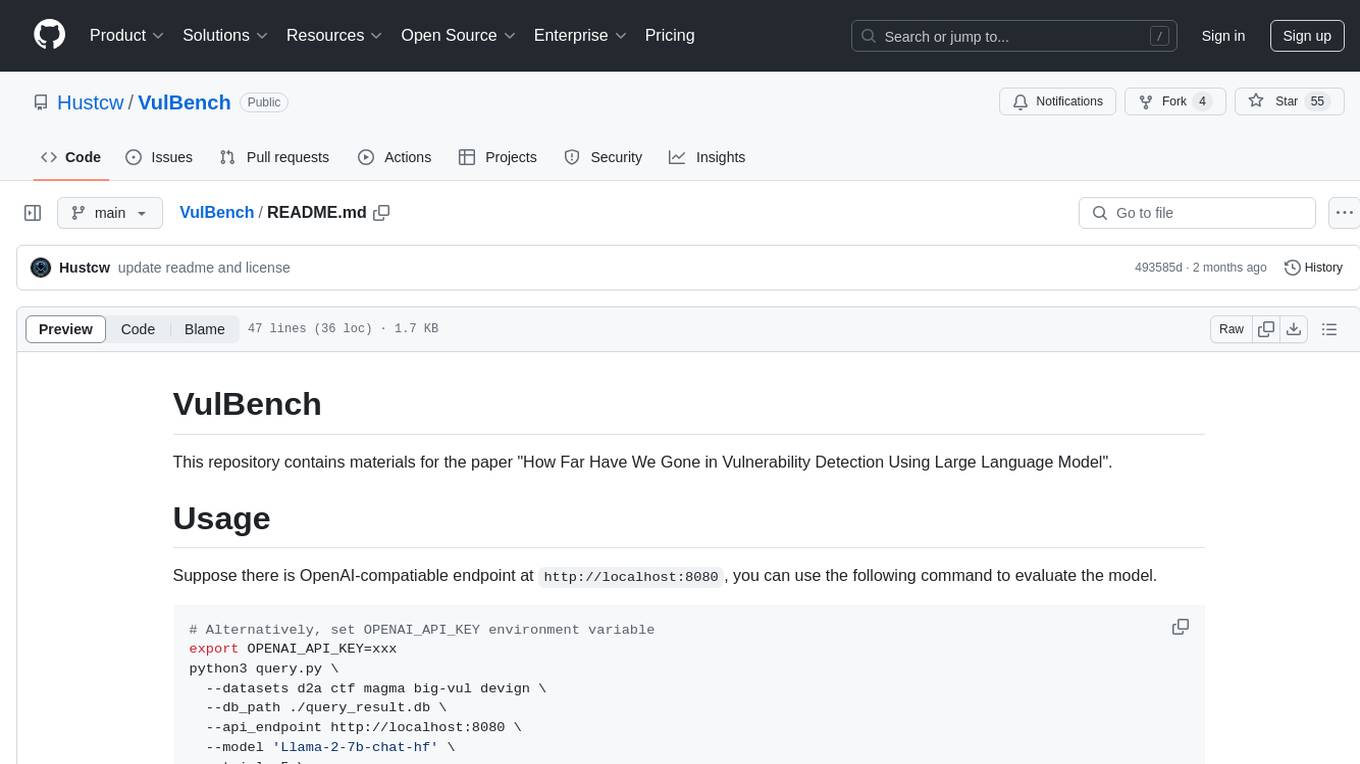
VulBench
This repository contains materials for the paper 'How Far Have We Gone in Vulnerability Detection Using Large Language Model'. It provides a tool for evaluating vulnerability detection models using datasets such as d2a, ctf, magma, big-vul, and devign. Users can query the model 'Llama-2-7b-chat-hf' and store results in a SQLite database for analysis. The tool supports binary and multiple classification tasks with concurrency settings. Additionally, users can evaluate the results and generate a CSV file with metrics for each dataset and prompt type.

verifAI
VerifAI is a document-based question-answering system that addresses hallucinations in generative large language models and search engines. It retrieves relevant documents, generates answers with references, and verifies answers for accuracy. The engine uses generative search technology and a verification model to ensure no misinformation. VerifAI supports various document formats and offers user registration with a React.js interface. It is open-source and designed to be user-friendly, making it accessible for anyone to use.
visualwebarena
VisualWebArena is a benchmark for evaluating multimodal autonomous language agents through diverse and complex web-based visual tasks. It builds on the reproducible evaluation introduced in WebArena. The repository provides scripts for end-to-end training, demos to run multimodal agents on webpages, and tools for setting up environments for evaluation. It includes trajectories of the GPT-4V + SoM agent on VWA tasks, along with human evaluations on 233 tasks. The environment supports OpenAI models and Gemini models for evaluation.
lotus
LOTUS (LLMs Over Tables of Unstructured and Structured Data) is a query engine that provides a declarative programming model and an optimized query engine for reasoning-based query pipelines over structured and unstructured data. It offers a simple and intuitive Pandas-like API with semantic operators for fast and easy LLM-powered data processing. The tool implements a semantic operator programming model, allowing users to write AI-based pipelines with high-level logic and leaving the rest of the work to the query engine. LOTUS supports various semantic operators like sem_map, sem_filter, sem_extract, sem_agg, sem_topk, sem_join, sem_sim_join, and sem_search, enabling users to perform tasks like mapping records, filtering data, aggregating records, and more. The tool also supports different model classes such as LM, RM, and Reranker for language modeling, retrieval, and reranking tasks respectively.
kaito
Kaito is an operator that automates the AI/ML inference model deployment in a Kubernetes cluster. It manages large model files using container images, avoids tuning deployment parameters to fit GPU hardware by providing preset configurations, auto-provisions GPU nodes based on model requirements, and hosts large model images in the public Microsoft Container Registry (MCR) if the license allows. Using Kaito, the workflow of onboarding large AI inference models in Kubernetes is largely simplified.
eval-dev-quality
DevQualityEval is an evaluation benchmark and framework designed to compare and improve the quality of code generation of Language Model Models (LLMs). It provides developers with a standardized benchmark to enhance real-world usage in software development and offers users metrics and comparisons to assess the usefulness of LLMs for their tasks. The tool evaluates LLMs' performance in solving software development tasks and measures the quality of their results through a point-based system. Users can run specific tasks, such as test generation, across different programming languages to evaluate LLMs' language understanding and code generation capabilities.
For similar tasks
phospho
Phospho is a text analytics platform for LLM apps. It helps you detect issues and extract insights from text messages of your users or your app. You can gather user feedback, measure success, and iterate on your app to create the best conversational experience for your users.
OpenFactVerification
Loki is an open-source tool designed to automate the process of verifying the factuality of information. It provides a comprehensive pipeline for dissecting long texts into individual claims, assessing their worthiness for verification, generating queries for evidence search, crawling for evidence, and ultimately verifying the claims. This tool is especially useful for journalists, researchers, and anyone interested in the factuality of information.
open-parse
Open Parse is a Python library for visually discerning document layouts and chunking them effectively. It is designed to fill the gap in open-source libraries for handling complex documents. Unlike text splitting, which converts a file to raw text and slices it up, Open Parse visually analyzes documents for superior LLM input. It also supports basic markdown for parsing headings, bold, and italics, and has high-precision table support, extracting tables into clean Markdown formats with accuracy that surpasses traditional tools. Open Parse is extensible, allowing users to easily implement their own post-processing steps. It is also intuitive, with great editor support and completion everywhere, making it easy to use and learn.
spaCy
spaCy is an industrial-strength Natural Language Processing (NLP) library in Python and Cython. It incorporates the latest research and is designed for real-world applications. The library offers pretrained pipelines supporting 70+ languages, with advanced neural network models for tasks such as tagging, parsing, named entity recognition, and text classification. It also facilitates multi-task learning with pretrained transformers like BERT, along with a production-ready training system and streamlined model packaging, deployment, and workflow management. spaCy is commercial open-source software released under the MIT license.
NanoLLM
NanoLLM is a tool designed for optimized local inference for Large Language Models (LLMs) using HuggingFace-like APIs. It supports quantization, vision/language models, multimodal agents, speech, vector DB, and RAG. The tool aims to provide efficient and effective processing for LLMs on local devices, enhancing performance and usability for various AI applications.

ontogpt
OntoGPT is a Python package for extracting structured information from text using large language models, instruction prompts, and ontology-based grounding. It provides a command line interface and a minimal web app for easy usage. The tool has been evaluated on test data and is used in related projects like TALISMAN for gene set analysis. OntoGPT enables users to extract information from text by specifying relevant terms and provides the extracted objects as output.

lima
LIMA is a multilingual linguistic analyzer developed by the CEA LIST, LASTI laboratory. It is Free Software available under the MIT license. LIMA has state-of-the-art performance for more than 60 languages using deep learning modules. It also includes a powerful rules-based mechanism called ModEx for extracting information in new domains without annotated data.
liboai
liboai is a simple C++17 library for the OpenAI API, providing developers with access to OpenAI endpoints through a collection of methods and classes. It serves as a spiritual port of OpenAI's Python library, 'openai', with similar structure and features. The library supports various functionalities such as ChatGPT, Audio, Azure, Functions, Image DALL·E, Models, Completions, Edit, Embeddings, Files, Fine-tunes, Moderation, and Asynchronous Support. Users can easily integrate the library into their C++ projects to interact with OpenAI services.
For similar jobs
sweep
Sweep is an AI junior developer that turns bugs and feature requests into code changes. It automatically handles developer experience improvements like adding type hints and improving test coverage.
teams-ai
The Teams AI Library is a software development kit (SDK) that helps developers create bots that can interact with Teams and Microsoft 365 applications. It is built on top of the Bot Framework SDK and simplifies the process of developing bots that interact with Teams' artificial intelligence capabilities. The SDK is available for JavaScript/TypeScript, .NET, and Python.
ai-guide
This guide is dedicated to Large Language Models (LLMs) that you can run on your home computer. It assumes your PC is a lower-end, non-gaming setup.

classifai
Supercharge WordPress Content Workflows and Engagement with Artificial Intelligence. Tap into leading cloud-based services like OpenAI, Microsoft Azure AI, Google Gemini and IBM Watson to augment your WordPress-powered websites. Publish content faster while improving SEO performance and increasing audience engagement. ClassifAI integrates Artificial Intelligence and Machine Learning technologies to lighten your workload and eliminate tedious tasks, giving you more time to create original content that matters.
chatbot-ui
Chatbot UI is an open-source AI chat app that allows users to create and deploy their own AI chatbots. It is easy to use and can be customized to fit any need. Chatbot UI is perfect for businesses, developers, and anyone who wants to create a chatbot.
BricksLLM
BricksLLM is a cloud native AI gateway written in Go. Currently, it provides native support for OpenAI, Anthropic, Azure OpenAI and vLLM. BricksLLM aims to provide enterprise level infrastructure that can power any LLM production use cases. Here are some use cases for BricksLLM: * Set LLM usage limits for users on different pricing tiers * Track LLM usage on a per user and per organization basis * Block or redact requests containing PIIs * Improve LLM reliability with failovers, retries and caching * Distribute API keys with rate limits and cost limits for internal development/production use cases * Distribute API keys with rate limits and cost limits for students
uAgents
uAgents is a Python library developed by Fetch.ai that allows for the creation of autonomous AI agents. These agents can perform various tasks on a schedule or take action on various events. uAgents are easy to create and manage, and they are connected to a fast-growing network of other uAgents. They are also secure, with cryptographically secured messages and wallets.
griptape
Griptape is a modular Python framework for building AI-powered applications that securely connect to your enterprise data and APIs. It offers developers the ability to maintain control and flexibility at every step. Griptape's core components include Structures (Agents, Pipelines, and Workflows), Tasks, Tools, Memory (Conversation Memory, Task Memory, and Meta Memory), Drivers (Prompt and Embedding Drivers, Vector Store Drivers, Image Generation Drivers, Image Query Drivers, SQL Drivers, Web Scraper Drivers, and Conversation Memory Drivers), Engines (Query Engines, Extraction Engines, Summary Engines, Image Generation Engines, and Image Query Engines), and additional components (Rulesets, Loaders, Artifacts, Chunkers, and Tokenizers). Griptape enables developers to create AI-powered applications with ease and efficiency.

