
matsciml
Open MatSci ML Toolkit is a framework for prototyping and scaling out deep learning models for materials discovery supporting widely used materials science datasets, and built on top of PyTorch Lightning, the Deep Graph Library, and PyTorch Geometric.
Stars: 170
The Open MatSci ML Toolkit is a flexible framework for machine learning in materials science. It provides a unified interface to a variety of materials science datasets, as well as a set of tools for data preprocessing, model training, and evaluation. The toolkit is designed to be easy to use for both beginners and experienced researchers, and it can be used to train models for a wide range of tasks, including property prediction, materials discovery, and materials design.
README:
This is the implementation of the MatSci ML benchmark, which includes ~1.5 million ground-state materials collected from various datasets, as well as integration of the OpenCatalyst dataset supporting diverse data format (point cloud, DGL graphs, PyG graphs), learning methods (single task, multi-task, multi-data) and deep learning models. Primary project contributors include: Santiago Miret (Intel Labs), Kin Long Kelvin Lee (Intel AXG), Carmelo Gonzales (Intel Labs), Mikhail Galkin (Intel Labs), Marcel Nassar (Intel Labs), Matthew Spellings (Vector Institute).
- [2024/08/23] Readthedocs is now online!
- [2023/09/27] Release of pre-packaged lmdb-based datasets from v1.0.0 via Zenodo.
- [2023/08/31] Initial release of the MatSci ML Benchmark with integration of ~1.5 million ground state materials.
- [2023/07/31] The Open MatSci ML Toolkit : A Flexible Framework for Deep Learning on the OpenCatalyst Dataset paper is accepted into TMLR. See previous version for code related to the benchmark.
The MatSci ML Benchmark contains diverse sets of tasks (energy prediction, force prediction, property prediction) across a broad range of datasets (OpenCatalyst Project [1], Materials Project [2], LiPS [3], OQMD [4], NOMAD [5], Carolina Materials Database [6]). Most of the data is related to energy prediction task, which is the most common property tracked for most materials systems in the literature. The codebase support single-task learning, as well as multi-task (training one model for multiple tasks within a dataset) and multi-date (training a model across multiple datsets with a common property). Additionally, we provide a generative materials pipeline that applies diffusion models (CDVAE [7]) to generate new unit cells.
The package follows the original design principles of the Open MatSci ML Toolkit, including:
- Ease of use for new ML researchers and practitioners that want get started on interacting with the OpenCatalyst dataset.
- Scalable computation of experiments leveraging PyTorch Lightning across different computation capabilities (laptop, server, cluster) and hardware platforms (CPU, GPU, XPU) without sacrificing performance in the compute and modeling.
- Integrating support for DGL and PyTorch Geometric for rapid GNN development.
The examples outlined in the next section how to get started with Open MatSci ML Toolkit using simple Python scripts, Jupyter notebooks, or the PyTorch Lightning CLI for a simple training on a portable subset of the original dataset (dev-set) that can be run on a laptop. Subsequently, we scale our example python script to large compute systems, including distributed data parallel training (multiple GPU on a single node) and multi-node training (multiple GPUs across multiple nodes) in a computing cluster. Leveraging both PyTorch Lightning and DGL capabilities, we can enable the compute and experiment scaling with minimal additional complexity.
-
Docker: We provide a Dockerfile inside thedockerthat can be run to install a container using standard docker commands. -
mamba: We have included amambaspecification that provides a complete out-of-the-box installation. Runmamba env create -n matsciml --file conda.yml, and will install all dependencies andmatscimlas an editable install. -
pip: In this case, we assume you are bringing your own virtual environment. Depending on what hardware platform you have, you can copy-paste the following commands; because the absolute mess that is modern Python packaging, these commands include the URLs for binary distributions of PyG and DGL graph backends.
For CPU only (good for local laptop development):
pip install -f https://data.pyg.org/whl/torch-2.4.0+cpu.html -f https://data.dgl.ai/wheels/torch-2.4/repo.html -e './[all]'For XPU usage, you will need to install PyTorch separately first, followed by matsciml; note that the PyTorch version is lower
as 2.3.1 is the latest XPU binary distributed.
pip install torch==2.3.1+cxx11.abi torchvision==0.18.1+cxx11.abi torchaudio==2.3.1+cxx11.abi intel-extension-for-pytorch==2.3.110+xpu oneccl_bind_pt==2.3.100+xpu --extra-index-url https://pytorch-extension.intel.com/release-whl/stable/xpu/us/
pip install -f https://data.pyg.org/whl/torch-2.3.0+cpu.html -f https://data.dgl.ai/wheels/torch-2.3/repo.html -e './[all]'For CUDA usage, substitute the index links with your particular toolkit version (e.g. 12.1 below):
pip install -f https://data.dgl.ai/wheels/torch-2.4/cu121/repo.html -f https://data.pyg.org/whl/torch-2.4.0+cu121.html -e './[all]'Additionally, for a development install, one can specify the extra packages like black and pytest with pip install './[dev]'. These can be
added to the commit workflow by running pre-commit install to generate git hooks.
[!NOTE] As of PyTorch 2.4+, XPU support has been upstreamed to PyTorch and starting from
torch>=2.5.0onwards, should be available as apipinstall. We will update the instructions accordingly when it does. We recommend consulting the PyTorch documentation for updates and instructions on how to get started with XPU use. In the meantime, please consult this page to see how to set up PyTorch on XPUs.
The module matsciml.lightning.xpu implements interfaces for Intel XPU to Lightning abstractions, including
the XPUAccelerator and two strategies for deployment (single XPU/tile and distributed data parallel).
Because we use PyTorch Lightning, there aren't many marked differences in running on Intel XPU, or GPUs
from other vendors. The abstractions we mentioned are registered in the various Lightning registries,
and should be accessible simply through pl.Trainer arguments, e.g.:
trainer = pl.Trainer(accelerator='xpu')The one major difference is for distributed data parallelism: Intel XPUs use the oneCCL communication
backend, which replaces nccl, gloo, or other backends typically passed to torch.distributed.
Please see examples/devices for single XPU/tile and DDP use cases.
NOTE: Currently there is a hard-coded torch.cuda.stream context in PyTorch Lightning's DDPStrategy.
This issue has been created to see if the maintainers would be happy to patch
it so that the cuda.Stream context is only used if a CUDA device is being used. If you encounter
a RuntimeError: Tried to instantiate dummy base class Stream, please just set ctx = nullcontext()
in the line of code that raises the exception.
The examples folder contains simple, unit scripts that demonstrate how to use the pipeline in specific ways:
Get started with different datasets with "devsets"
# Materials project
python examples/datasets/materials_project/single_task_devset.py
# Carolina materials database
python examples/datasets/carolina_db/single_task_devset.py
# NOMAD
python examples/datasets/nomad/single_task_devset.py
# OQMD
python examples/datasets/oqmd/single_task_devset.pyRepresentation learning with symmetry pretraining
# uses the devset for synthetic point group point clouds
python examples/tasks/symmetry/single_symmetry_example.pyExample notebook-based development and testing
jupyter notebook examples/devel-example.ipynbFor more advanced use cases:
Checkout materials generation with CDVAE
CDVAE [7] is a latent diffusion model that trains a VAE on the reconstruction objective, adds Gaussian noise to the latent variable, and learns to predict the noise. The noised and generated features inlcude lattice parameters, atoms composition, and atom coordinates. The generation process is based on the annealed Langevin dynamics.
CDVAE is implemented in the GenerationTask and we provide a custom data
split from the Materials Project bounded by 25 atoms per structure.
The process is split into 3 parts with 3 respective scripts found in
examples/model_demos/cdvae/.
- Training CDVAE on the reconstruction and denoising objectives:
cdvae.py - Sampling the structures (from scratch or reconstruct the test set):
cdvae_inference.py - Evaluating the sampled structures:
cdvae_metrics.py
The sampling procedure takes some time (about 5-8 hours for 10000 structures
depending on the hardware) due to the Langevin dynamics.
The default hyperparameters of CDVAE components correspond to that from the
original paper and can be found in cdvae_configs.py.
# training
python examples/model_demos/cdvae/cdvae.py --data_path <path/to/splits>
# sampling 10,000 structures from scratch
python examples/model_demos/cdvae/cdvae_inference.py --model_path <path/to/checkpoint> --data_path <path/to/splits> --tasks gen
# evaluating the sampled structures
python examples/model_demos/cdvae/cdvae_metrics.py --root_path <path/to/generated_samples> --data_path <path/to/splits> --tasks genMultiple tasks trained using the same dataset
# this script requires modification as you'll need to download the materials
# project dataset, and point L24 to the folder where it was saved
python examples/tasks/multitask/single_data_multitask_example.pyUtilizes Materials Project data to train property regression and material classification jointly
Multiple tasks trained using multiple datasets
python examples/tasks/multitask/three_datasets.pyTrain regression tasks against IS2RE, S2EF, and LiPS datasets jointly
In the scripts folder you will find two scripts needed to download and preprocess datasets: the download_datasets.py can be used to obtain Carolina DB, Materials Project, NOMAD, and OQMD datasets, while the download_ocp_data.py preserves the original Open Catalyst script.
In the current release, we have implemented interfaces to a number of large scale materials science datasets. Under the hood, the data structures pulled from each dataset have been homogenized, and the only real interaction layer for users is through the MatSciMLDataModule, a subclass of LightningDataModule.
from matsciml.lightning.data_utils import MatSciMLDataModule
# no configuration needed, although one can specify the batch size and number of workers
devset_module = MatSciMLDataModule.from_devset(dataset="MaterialsProjectDataset")This will let you springboard into development without needing to worry about how to wrangle with the datasets; just grab a batch and go! With the exception of Open Catalyst, datasets will typically return point cloud representations; we provide a flexible transform interface to interconvert between representations and frameworks:
From point clouds to DGL graphs
from matsciml.datasets.transforms import PointCloudToGraphTransform
# make the materials project dataset emit DGL graphs, based on a atom-atom distance cutoff of 10
devset = MatSciMLDataModule.from_devset(
dataset="MaterialsProjectDataset",
dset_kwargs={"transforms": [PointCloudToGraphTransform(backend="dgl", cutoff_dist=10.)]}
)But I want to use PyG?
from matsciml.datasets.transforms import PointCloudToGraphTransform
# change the backend argument to obtain PyG graphs
devset = MatSciMLDataModule.from_devset(
dataset="MaterialsProjectDataset",
dset_kwargs={"transforms": [PointCloudToGraphTransform(backend="pyg", cutoff_dist=10.)]}
)What else can I configure with `MatSciMLDataModule`?
Datasets beyond devsets can be configured through class arguments:
devset = MatSciMLDataModule(
dataset="MaterialsProjectDataset",
train_path="/path/to/training/lmdb/folder",
batch_size=64,
num_workers=4, # configure data loader instances
dset_kwargs={"transforms": [PointCloudToGraphTransform(backend="pyg", cutoff_dist=10.)]},
val_split="/path/to/val/lmdb/folder"
)In particular, val_split and test_split can point to their LMDB folders, or just a float between [0,1] to do quick, uniform splits. The rest, including distributed sampling, will be taken care of for you under the hood.
How do I compose multiple datasets?
Given the amount of configuration involved, composing multiple datasets takes a little more work but we have tried to make it as seamless as possible. The main difference from the single dataset case is replacing MatSciMLDataModule with MultiDataModule from matsciml.lightning.data_utils, configuring each dataset manually, and passing them collectively into the data module:
from matsciml.datasets import MaterialsProjectDataset, OQMDDataset, MultiDataset
from matsciml.lightning.data_utils import MultiDataModule
# configure training only here, but same logic extends to validation/test splits
train_dset = MultiDataset(
[
MaterialsProjectDataset("/path/to/train/materialsproject"),
OQMDDataset("/path/to/train/oqmd")
]
)
# this configures the actual data module passed into Lightning
datamodule = MultiDataModule(
batch_size=32,
num_workers=4,
train_dataset=train_dset
)While it does require a bit of extra work, this was to ensure flexibility in how you can compose datasets. We welcome feedback on the user experience! 😃
In Open MatSci ML Toolkit, tasks effective form learning objectives: at a high level, a task takes an encoding model/backbone that ingests a structure to predict one or several properties, or classify a material. In the single task case, there may be multiple targets and the neural network architecture may be fluid, but there is only one optimizer. Under this definition, multi-task learning comprises multiple tasks and optimizers operating jointly through a single embedding.
- [1] Chanussot, L., Das, A., Goyal, S., Lavril, T., Shuaibi, M., Riviere, M., Tran, K., Heras-Domingo, J., Ho, C., Hu, W. and Palizhati, A., 2021. Open catalyst 2020 (OC20) dataset and community challenges. Acs Catalysis, 11(10), pp.6059-6072.
- [2] Jain, A., Ong, S.P., Hautier, G., Chen, W., Richards, W.D., Dacek, S., Cholia, S., Gunter, D., Skinner, D., Ceder, G. and Persson, K.A., 2013. Commentary: The Materials Project: A materials genome approach to accelerating materials innovation. APL materials, 1(1).
- [3] Batzner, S., Musaelian, A., Sun, L., Geiger, M., Mailoa, J.P., Kornbluth, M., Molinari, N., Smidt, T.E. and Kozinsky, B., 2022. E (3)-equivariant graph neural networks for data-efficient and accurate interatomic potentials. Nature communications, 13(1), p.2453.
- [4] Kirklin, S., Saal, J.E., Meredig, B., Thompson, A., Doak, J.W., Aykol, M., Rühl, S. and Wolverton, C., 2015. The Open Quantum Materials Database (OQMD): assessing the accuracy of DFT formation energies. npj Computational Materials, 1(1), pp.1-15.
- [5] Draxl, C. and Scheffler, M., 2019. The NOMAD laboratory: from data sharing to artificial intelligence. Journal of Physics: Materials, 2(3), p.036001.
- [6] Zhao, Y., Al‐Fahdi, M., Hu, M., Siriwardane, E.M., Song, Y., Nasiri, A. and Hu, J., 2021. High‐throughput discovery of novel cubic crystal materials using deep generative neural networks. Advanced Science, 8(20), p.2100566.
- [7] Xie, T., Fu, X., Ganea, O.E., Barzilay, R. and Jaakkola, T.S., 2021, October. Crystal Diffusion Variational Autoencoder for Periodic Material Generation. In International Conference on Learning Representations.
Please refer to the developers guide for how to contribute the the Open MatSciML Toolkit.
If you use Open MatSci ML Toolkit in your technical work or publication, we would appreciate it if you cite the Open MatSci ML Toolkit paper in TMLR:
Miret, S.; Lee, K. L. K.; Gonzales, C.; Nassar, M.; Spellings, M. The Open MatSci ML Toolkit: A Flexible Framework for Machine Learning in Materials Science. Transactions on Machine Learning Research, 2023.
@article{openmatscimltoolkit,
title = {The Open {{MatSci ML}} Toolkit: {{A}} Flexible Framework for Machine Learning in Materials Science},
author = {Miret, Santiago and Lee, Kin Long Kelvin and Gonzales, Carmelo and Nassar, Marcel and Spellings, Matthew},
year = {2023},
journal = {Transactions on Machine Learning Research},
issn = {2835-8856}
}If you use v1.0.0, please cite our paper:
Lee, K. L. K., Gonzales, C., Nassar, M., Spellings, M., Galkin, M., & Miret, S. (2023). MatSciML: A Broad, Multi-Task Benchmark for Solid-State Materials Modeling. arXiv preprint arXiv:2309.05934.
@article{lee2023matsciml,
title={MatSciML: A Broad, Multi-Task Benchmark for Solid-State Materials Modeling},
author={Lee, Kin Long Kelvin and Gonzales, Carmelo and Nassar, Marcel and Spellings, Matthew and Galkin, Mikhail and Miret, Santiago},
journal={arXiv preprint arXiv:2309.05934},
year={2023}
}Please cite datasets used in your work as well. You can find additional descriptions and details regarding each dataset here.
For Tasks:
Click tags to check more tools for each tasksFor Jobs:
Alternative AI tools for matsciml
Similar Open Source Tools

matsciml
The Open MatSci ML Toolkit is a flexible framework for machine learning in materials science. It provides a unified interface to a variety of materials science datasets, as well as a set of tools for data preprocessing, model training, and evaluation. The toolkit is designed to be easy to use for both beginners and experienced researchers, and it can be used to train models for a wide range of tasks, including property prediction, materials discovery, and materials design.
AIF360
The AI Fairness 360 toolkit is an open-source library designed to detect and mitigate bias in machine learning models. It provides a comprehensive set of metrics, explanations, and algorithms for bias mitigation in various domains such as finance, healthcare, and education. The toolkit supports multiple bias mitigation algorithms and fairness metrics, and is available in both Python and R. Users can leverage the toolkit to ensure fairness in AI applications and contribute to its development for extensibility.
torchtitan
Torchtitan is a PyTorch native platform designed for rapid experimentation and large-scale training of generative AI models. It provides a flexible foundation for developers to build upon with extension points for creating custom extensions. The tool showcases PyTorch's latest distributed training features and supports pretraining Llama 3.1 LLMs of various sizes. It offers key features like multi-dimensional parallelisms, FSDP2 with per-parameter sharding, Tensor Parallel, Pipeline Parallel, and more. Users can contribute to the tool through the experiments folder and core contributions guidelines. Installation can be done from source, nightly builds, or stable releases. The tool also supports training Llama 3.1 models and provides guidance on starting a training run and multi-node training. Citation information is available for referencing the tool in academic work, and the source code is under a BSD 3 license.
LLMeBench
LLMeBench is a flexible framework designed for accelerating benchmarking of Large Language Models (LLMs) in the field of Natural Language Processing (NLP). It supports evaluation of various NLP tasks using model providers like OpenAI, HuggingFace Inference API, and Petals. The framework is customizable for different NLP tasks, LLM models, and datasets across multiple languages. It features extensive caching capabilities, supports zero- and few-shot learning paradigms, and allows on-the-fly dataset download and caching. LLMeBench is open-source and continuously expanding to support new models accessible through APIs.

semlib
Semlib is a Python library for building data processing and data analysis pipelines that leverage the power of large language models (LLMs). It provides functional programming primitives like map, reduce, sort, and filter, programmed with natural language descriptions. Semlib handles complexities such as prompting, parsing, concurrency control, caching, and cost tracking. The library breaks down sophisticated data processing tasks into simpler steps to improve quality, feasibility, latency, cost, security, and flexibility of data processing tasks.
kafka-ml
Kafka-ML is a framework designed to manage the pipeline of Tensorflow/Keras and PyTorch machine learning models on Kubernetes. It enables the design, training, and inference of ML models with datasets fed through Apache Kafka, connecting them directly to data streams like those from IoT devices. The Web UI allows easy definition of ML models without external libraries, catering to both experts and non-experts in ML/AI.

chem-bench
ChemBench is a project aimed at expanding chemistry benchmark tasks in a BIG-bench compatible way, providing a pipeline to benchmark frontier and open models. It allows users to run benchmarking tasks on models with existing presets, offering predefined parameters and processing steps. The library facilitates benchmarking models on the entire suite, addressing challenges such as prompt structure, parsing, and scoring methods. Users can contribute to the project by following the developer notes.
codellm-devkit
Codellm-devkit (CLDK) is a Python library that serves as a multilingual program analysis framework bridging traditional static analysis tools and Large Language Models (LLMs) specialized for code (CodeLLMs). It simplifies the process of analyzing codebases across multiple programming languages, enabling the extraction of meaningful insights and facilitating LLM-based code analysis. The library provides a unified interface for integrating outputs from various analysis tools and preparing them for effective use by CodeLLMs. Codellm-devkit aims to enable the development and experimentation of robust analysis pipelines that combine traditional program analysis tools and CodeLLMs, reducing friction in multi-language code analysis and ensuring compatibility across different tools and LLM platforms. It is designed to seamlessly integrate with popular analysis tools like WALA, Tree-sitter, LLVM, and CodeQL, acting as a crucial intermediary layer for efficient communication between these tools and CodeLLMs. The project is continuously evolving to include new tools and frameworks, maintaining its versatility for code analysis and LLM integration.
lotus
LOTUS (LLMs Over Tables of Unstructured and Structured Data) is a query engine that provides a declarative programming model and an optimized query engine for reasoning-based query pipelines over structured and unstructured data. It offers a simple and intuitive Pandas-like API with semantic operators for fast and easy LLM-powered data processing. The tool implements a semantic operator programming model, allowing users to write AI-based pipelines with high-level logic and leaving the rest of the work to the query engine. LOTUS supports various semantic operators like sem_map, sem_filter, sem_extract, sem_agg, sem_topk, sem_join, sem_sim_join, and sem_search, enabling users to perform tasks like mapping records, filtering data, aggregating records, and more. The tool also supports different model classes such as LM, RM, and Reranker for language modeling, retrieval, and reranking tasks respectively.

zshot
Zshot is a highly customizable framework for performing Zero and Few shot named entity and relationships recognition. It can be used for mentions extraction, wikification, zero and few shot named entity recognition, zero and few shot named relationship recognition, and visualization of zero-shot NER and RE extraction. The framework consists of two main components: the mentions extractor and the linker. There are multiple mentions extractors and linkers available, each serving a specific purpose. Zshot also includes a relations extractor and a knowledge extractor for extracting relations among entities and performing entity classification. The tool requires Python 3.6+ and dependencies like spacy, torch, transformers, evaluate, and datasets for evaluation over datasets like OntoNotes. Optional dependencies include flair and blink for additional functionalities. Zshot provides examples, tutorials, and evaluation methods to assess the performance of the components.
fuse-med-ml
FuseMedML is a Python framework designed to accelerate machine learning-based discovery in the medical field by promoting code reuse. It provides a flexible design concept where data is stored in a nested dictionary, allowing easy handling of multi-modality information. The framework includes components for creating custom models, loss functions, metrics, and data processing operators. Additionally, FuseMedML offers 'batteries included' key components such as fuse.data for data processing, fuse.eval for model evaluation, and fuse.dl for reusable deep learning components. It supports PyTorch and PyTorch Lightning libraries and encourages the creation of domain extensions for specific medical domains.
storm
STORM is a LLM system that writes Wikipedia-like articles from scratch based on Internet search. While the system cannot produce publication-ready articles that often require a significant number of edits, experienced Wikipedia editors have found it helpful in their pre-writing stage. **Try out our [live research preview](https://storm.genie.stanford.edu/) to see how STORM can help your knowledge exploration journey and please provide feedback to help us improve the system 🙏!**
baal
Baal is an active learning library that supports both industrial applications and research use cases. It provides a framework for Bayesian active learning methods such as Monte-Carlo Dropout, MCDropConnect, Deep ensembles, and Semi-supervised learning. Baal helps in labeling the most uncertain items in the dataset pool to improve model performance and reduce annotation effort. The library is actively maintained by a dedicated team and has been used in various research papers for production and experimentation.
cuvs
cuVS is a library that contains state-of-the-art implementations of several algorithms for running approximate nearest neighbors and clustering on the GPU. It can be used directly or through the various databases and other libraries that have integrated it. The primary goal of cuVS is to simplify the use of GPUs for vector similarity search and clustering.
llm-analysis
llm-analysis is a tool designed for Latency and Memory Analysis of Transformer Models for Training and Inference. It automates the calculation of training or inference latency and memory usage for Large Language Models (LLMs) or Transformers based on specified model, GPU, data type, and parallelism configurations. The tool helps users to experiment with different setups theoretically, understand system performance, and optimize training/inference scenarios. It supports various parallelism schemes, communication methods, activation recomputation options, data types, and fine-tuning strategies. Users can integrate llm-analysis in their code using the `LLMAnalysis` class or use the provided entry point functions for command line interface. The tool provides lower-bound estimations of memory usage and latency, and aims to assist in achieving feasible and optimal setups for training or inference.
nitrain
Nitrain is a framework for medical imaging AI that provides tools for sampling and augmenting medical images, training models on medical imaging datasets, and visualizing model results in a medical imaging context. It supports using pytorch, keras, and tensorflow.
For similar tasks

matsciml
The Open MatSci ML Toolkit is a flexible framework for machine learning in materials science. It provides a unified interface to a variety of materials science datasets, as well as a set of tools for data preprocessing, model training, and evaluation. The toolkit is designed to be easy to use for both beginners and experienced researchers, and it can be used to train models for a wide range of tasks, including property prediction, materials discovery, and materials design.
aideml
AIDE is a machine learning code generation agent that can generate solutions for machine learning tasks from natural language descriptions. It has the following features: 1. **Instruct with Natural Language**: Describe your problem or additional requirements and expert insights, all in natural language. 2. **Deliver Solution in Source Code**: AIDE will generate Python scripts for the **tested** machine learning pipeline. Enjoy full transparency, reproducibility, and the freedom to further improve the source code! 3. **Iterative Optimization**: AIDE iteratively runs, debugs, evaluates, and improves the ML code, all by itself. 4. **Visualization**: We also provide tools to visualize the solution tree produced by AIDE for a better understanding of its experimentation process. This gives you insights not only about what works but also what doesn't. AIDE has been benchmarked on over 60 Kaggle data science competitions and has demonstrated impressive performance, surpassing 50% of Kaggle participants on average. It is particularly well-suited for tasks that require complex data preprocessing, feature engineering, and model selection.

AgentCPM
AgentCPM is a series of open-source LLM agents jointly developed by THUNLP, Renmin University of China, ModelBest, and the OpenBMB community. It addresses challenges faced by agents in real-world applications such as limited long-horizon capability, autonomy, and generalization. The team focuses on building deep research capabilities for agents, releasing AgentCPM-Explore, a deep-search LLM agent, and AgentCPM-Report, a deep-research LLM agent. AgentCPM-Explore is the first open-source agent model with 4B parameters to appear on widely used long-horizon agent benchmarks. AgentCPM-Report is built on the 8B-parameter base model MiniCPM4.1, autonomously generating long-form reports with extreme performance and minimal footprint, designed for high-privacy scenarios with offline and agile local deployment.
For similar jobs

matsciml
The Open MatSci ML Toolkit is a flexible framework for machine learning in materials science. It provides a unified interface to a variety of materials science datasets, as well as a set of tools for data preprocessing, model training, and evaluation. The toolkit is designed to be easy to use for both beginners and experienced researchers, and it can be used to train models for a wide range of tasks, including property prediction, materials discovery, and materials design.

NoLabs
NoLabs is an open-source biolab that provides easy access to state-of-the-art models for bio research. It supports various tasks, including drug discovery, protein analysis, and small molecule design. NoLabs aims to accelerate bio research by making inference models accessible to everyone.

AlphaFold3
AlphaFold3 is an implementation of the Alpha Fold 3 model in PyTorch for accurate structure prediction of biomolecular interactions. It includes modules for genetic diffusion and full model examples for forward pass computations. The tool allows users to generate random pair and single representations, operate on atomic coordinates, and perform structure predictions based on input tensors. The implementation also provides functionalities for training and evaluating the model.
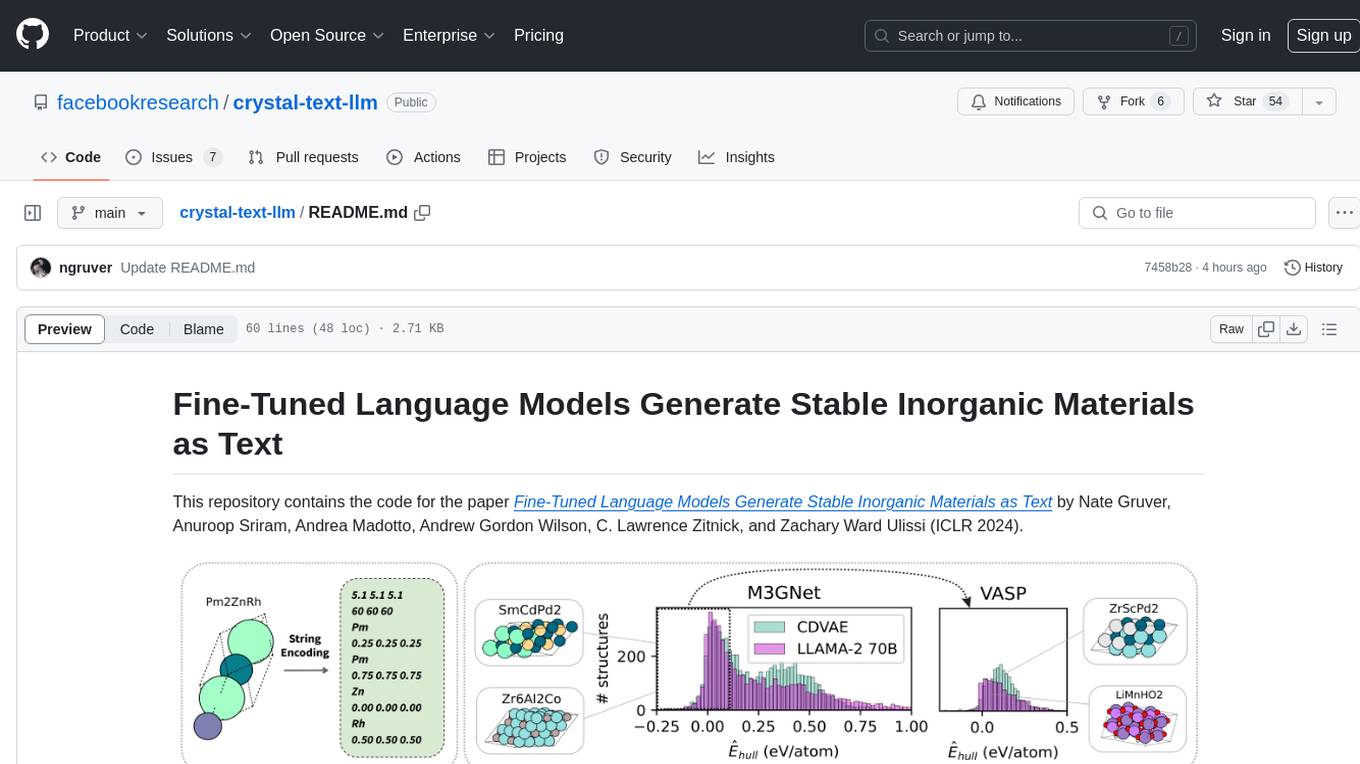
crystal-text-llm
This repository contains the code for the paper Fine-Tuned Language Models Generate Stable Inorganic Materials as Text. It demonstrates how finetuned LLMs can be used to generate stable materials, match or exceed the performance of domain specific models, mutate existing materials, and sample crystal structures conditioned on text descriptions. The method is distinct from CrystaLLM, which trains language models from scratch on CIF-formatted crystals.

Scientific-LLM-Survey
Scientific Large Language Models (Sci-LLMs) is a repository that collects papers on scientific large language models, focusing on biology and chemistry domains. It includes textual, molecular, protein, and genomic languages, as well as multimodal language. The repository covers various large language models for tasks such as molecule property prediction, interaction prediction, protein sequence representation, protein sequence generation/design, DNA-protein interaction prediction, and RNA prediction. It also provides datasets and benchmarks for evaluating these models. The repository aims to facilitate research and development in the field of scientific language modeling.
md-agent
MD-Agent is a LLM-agent based toolset for Molecular Dynamics. It uses Langchain and a collection of tools to set up and execute molecular dynamics simulations, particularly in OpenMM. The tool assists in environment setup, installation, and usage by providing detailed steps. It also requires API keys for certain functionalities, such as OpenAI and paper-qa for literature searches. Contributions to the project are welcome, with a detailed Contributor's Guide available for interested individuals.
AIMNet2
AIMNet2 Calculator is a package that integrates the AIMNet2 neural network potential into simulation workflows, providing fast and reliable energy, force, and property calculations for molecules with diverse elements. It excels at modeling various systems, offers flexible interfaces for popular simulation packages, and supports long-range interactions using DSF or Ewald summation Coulomb models. The tool is designed for accurate and versatile molecular simulations, suitable for large molecules and periodic calculations.
admet_ai
ADMET-AI is a platform for ADMET prediction using Chemprop-RDKit models trained on ADMET datasets from the Therapeutics Data Commons. It offers command line, Python API, and web server interfaces for making ADMET predictions on new molecules. The platform can be easily installed using pip and supports GPU acceleration. It also provides options for processing TDC data, plotting results, and hosting a web server. ADMET-AI is a machine learning platform for evaluating large-scale chemical libraries.







