LLM-for-genomics-training
Tutorial on large language models for genomics
Stars: 192
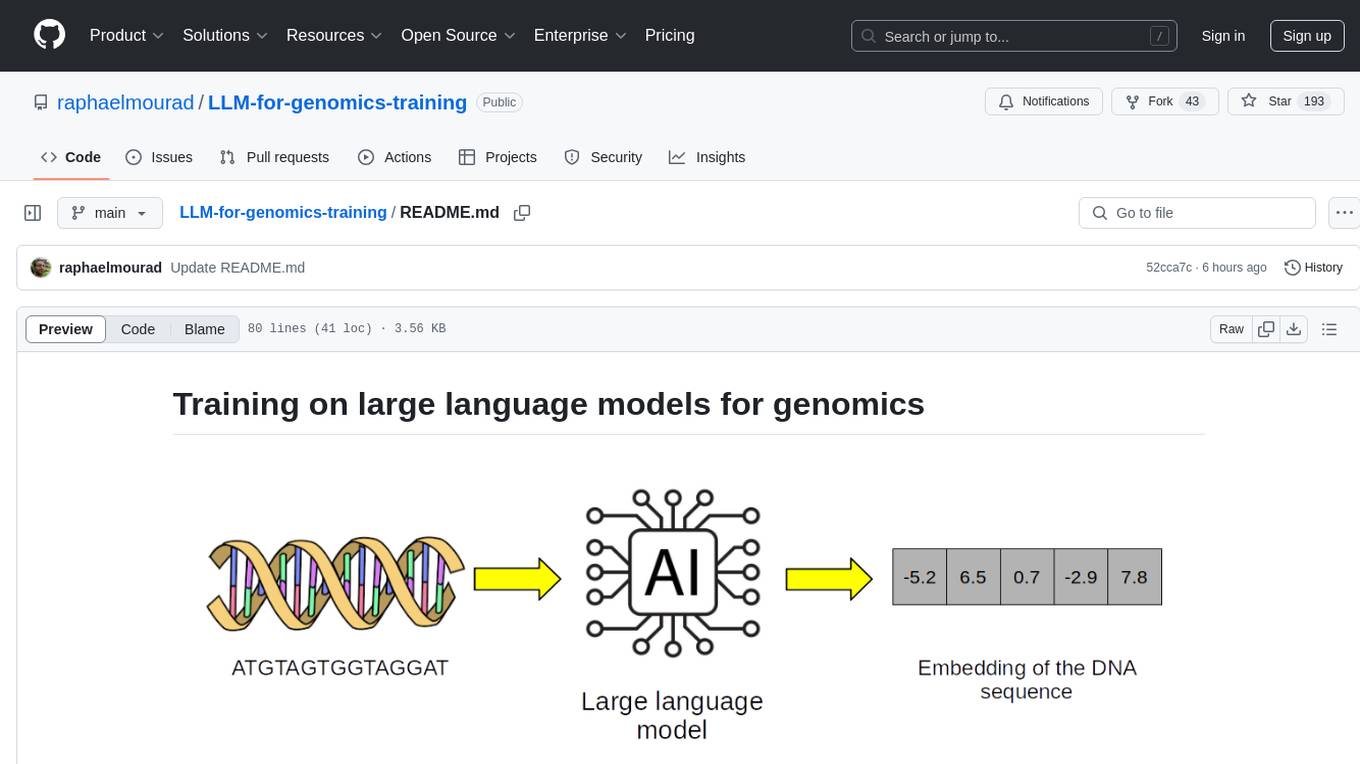
This repository provides training on large language models (LLMs) for genomics, including lecture notes and lab classes covering pretraining, finetuning, zeroshot learning prediction of mutation effect, synthetic DNA sequence generation, and DNA sequence optimization.
README:
In this repository, we will follow a training for large language models (LLMs) for genomics. The training comprises a short lecture and several lab classes.
You can download the lecture note here.
The data can be found in the file:
- data/genome_sequences/hg38/sequences_hg38_200b_verysmall.csv.gz
The file contains 100,000 non-overlapping DNA sequences of 200 bases, corresponding to around 1% of the human genome. For instance, here is one DNA sequence of 200 bases:

We will pretrain an LLM from scratch (a simplified mistral model, see folder data/models/Mixtral-8x7B-v0.1/) on the 100,000 DNA sequences from the human genome. The LLM is pretrained with causal language modeling using 200b DNA sequences from the human genome hg38 assembly.
We will use a pretrained LLM from huggingface (https://huggingface.co/RaphaelMourad/Mistral-DNA-v1-17M-hg38) and finetune it for DNA sequence classification. The aim is to classify a DNA sequence depending on whether it binds a protein or not (transcription factor), or if a histone mark is present, or if a promoter is active.
We will use a pretrained LLM from huggingface (https://huggingface.co/RaphaelMourad/Mistral-DNA-v1-17M-hg38) to predict the impact of mutations with zeroshot learning (directly using the pretrained model for DNA sequences). Here, we compute the embedding of the wild type sequence and compare it to the embedding of the mutated sequence, and then compute a L2 distance between the two embeddings. We expect that the higher the distance, the larger the mutation effect.
We will use a pretrained LLM from huggingface (https://huggingface.co/RaphaelMourad/Mistral-DNA-v1-138M-yeast) to generate artificial yeast DNA sequences.
We will use a finetuned LLM for promoter or transcription factor binding.
For Tasks:
Click tags to check more tools for each tasksFor Jobs:
Alternative AI tools for LLM-for-genomics-training
Similar Open Source Tools

LLM-for-genomics-training
This repository provides training on large language models (LLMs) for genomics, including lecture notes and lab classes covering pretraining, finetuning, zeroshot learning prediction of mutation effect, synthetic DNA sequence generation, and DNA sequence optimization.

DNAnalyzer
DNAnalyzer is a nonprofit organization dedicated to revolutionizing DNA analysis through AI-powered tools. It aims to democratize access to DNA analysis for a deeper understanding of human health and disease. The tool provides innovative AI-powered analysis and interpretive tools to empower geneticists, physicians, and researchers to gain deep insights into DNA sequences, revolutionizing how we understand human health and disease.
PINNACLE
PINNACLE is a flexible geometric deep learning approach that trains on contextualized protein interaction networks to generate context-aware protein representations. It provides protein representations split across various cell-type contexts from different tissues and organs. The tool can be fine-tuned to study the genomic effects of drugs and nominate promising protein targets and cell-type contexts for further investigation. PINNACLE exemplifies the paradigm of incorporating context-specific effects for studying biological systems, especially the impact of disease and therapeutics.
awesome-synthetic-datasets
This repository focuses on organizing resources for building synthetic datasets using large language models. It covers important datasets, libraries, tools, tutorials, and papers related to synthetic data generation. The goal is to provide pragmatic and practical resources for individuals interested in creating synthetic datasets for machine learning applications.
xlstm-jax
The xLSTM-jax repository contains code for training and evaluating the xLSTM model on language modeling using JAX. xLSTM is a Recurrent Neural Network architecture that improves upon the original LSTM through Exponential Gating, normalization, stabilization techniques, and a Matrix Memory. It is optimized for large-scale distributed systems with performant triton kernels for faster training and inference.
ManipVQA
ManipVQA is a framework that enhances Multimodal Large Language Models (MLLMs) with manipulation-centric knowledge through a Visual Question-Answering (VQA) format. It addresses the deficiency of conventional MLLMs in understanding affordances and physical concepts crucial for manipulation tasks. By infusing robotics-specific knowledge, including tool detection, affordance recognition, and physical concept comprehension, ManipVQA improves the performance of robots in manipulation tasks. The framework involves fine-tuning MLLMs with a curated dataset of interactive objects, enabling robots to understand and execute natural language instructions more effectively.
matchem-llm
A public repository collecting links to state-of-the-art training sets, QA, benchmarks and other evaluations for various ML and LLM applications in materials science and chemistry. It includes datasets related to chemistry, materials, multimodal data, and knowledge graphs in the field. The repository aims to provide resources for training and evaluating machine learning models in the materials science and chemistry domains.
Graph-CoT
This repository contains the source code and datasets for Graph Chain-of-Thought: Augmenting Large Language Models by Reasoning on Graphs accepted to ACL 2024. It proposes a framework called Graph Chain-of-thought (Graph-CoT) to enable Language Models to traverse graphs step-by-step for reasoning, interaction, and execution. The motivation is to alleviate hallucination issues in Language Models by augmenting them with structured knowledge sources represented as graphs.

ai_projects
This repository contains a collection of AI projects covering various areas of machine learning. Each project is accompanied by detailed articles on the associated blog sciblog. Projects range from introductory topics like Convolutional Neural Networks and Transfer Learning to advanced topics like Fraud Detection and Recommendation Systems. The repository also includes tutorials on data generation, distributed training, natural language processing, and time series forecasting. Additionally, it features visualization projects such as football match visualization using Datashader.
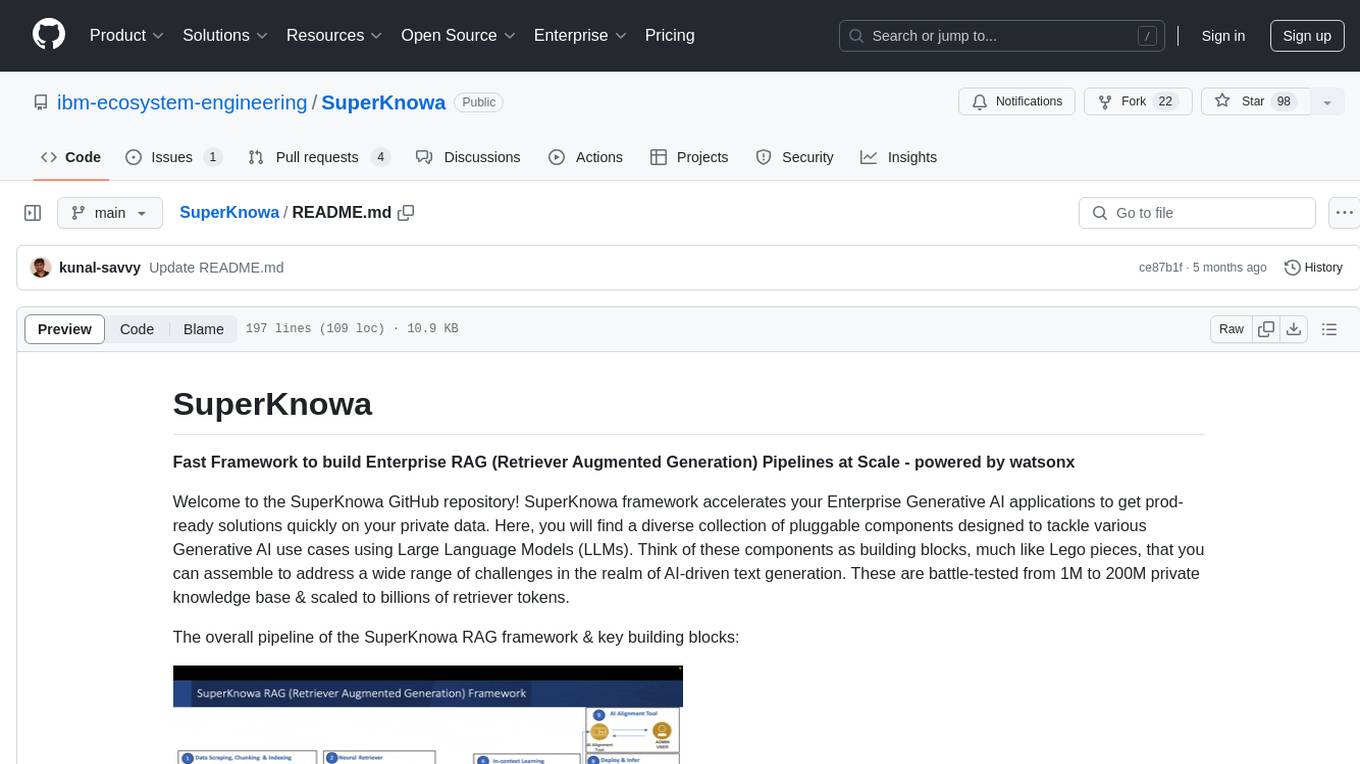
SuperKnowa
SuperKnowa is a fast framework to build Enterprise RAG (Retriever Augmented Generation) Pipelines at Scale, powered by watsonx. It accelerates Enterprise Generative AI applications to get prod-ready solutions quickly on private data. The framework provides pluggable components for tackling various Generative AI use cases using Large Language Models (LLMs), allowing users to assemble building blocks to address challenges in AI-driven text generation. SuperKnowa is battle-tested from 1M to 200M private knowledge base & scaled to billions of retriever tokens.
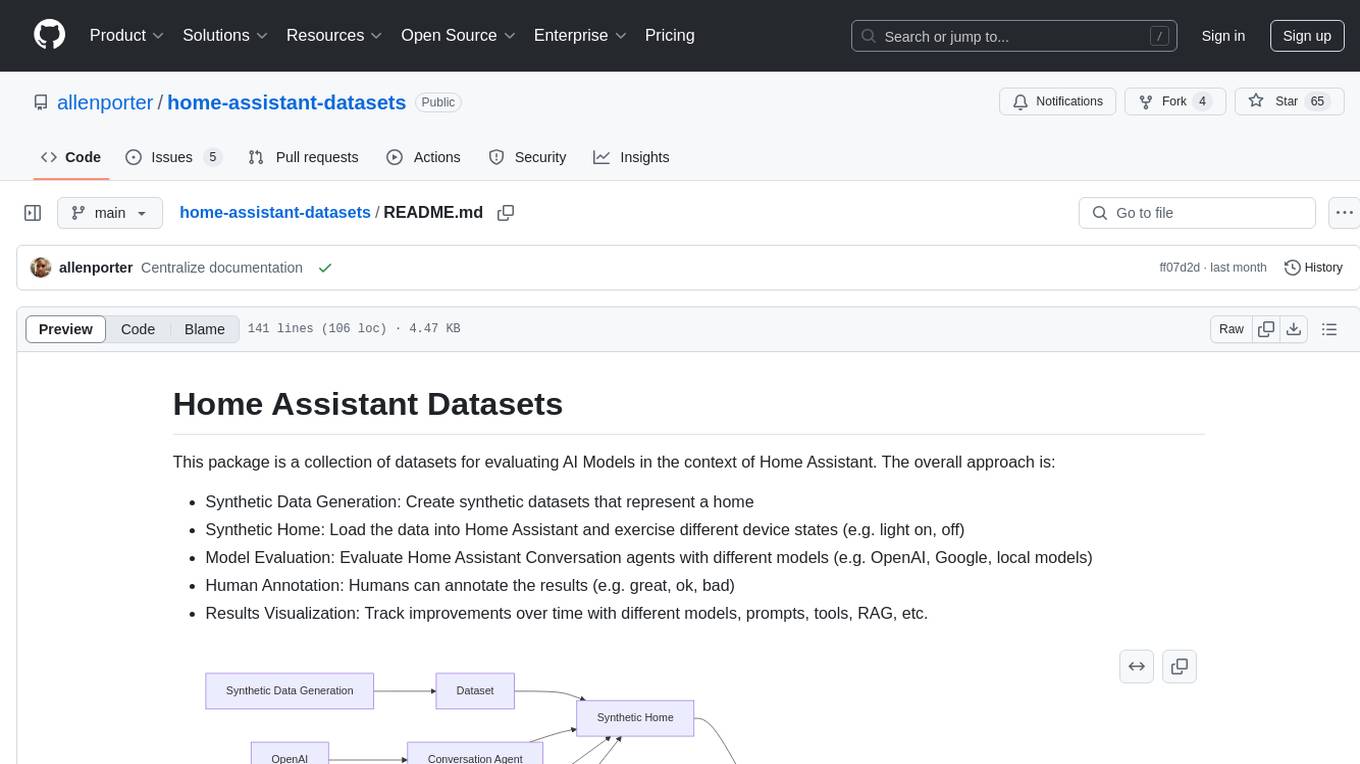
home-assistant-datasets
This package provides a collection of datasets for evaluating AI Models in the context of Home Assistant. It includes synthetic data generation, loading data into Home Assistant, model evaluation with different conversation agents, human annotation of results, and visualization of improvements over time. The datasets cover home descriptions, area descriptions, device descriptions, and summaries that can be performed on a home. The tool aims to build datasets for future training purposes.
raft
RAFT (Retrieval-Augmented Fine-Tuning) is a method for creating conversational agents that realistically emulate specific human targets. It involves a dual-phase process of fine-tuning and retrieval-based augmentation to generate nuanced and personalized dialogue. The tool is designed to combine interview transcripts with memories from past writings to enhance language model responses. RAFT has the potential to advance the field of personalized, context-sensitive conversational agents.
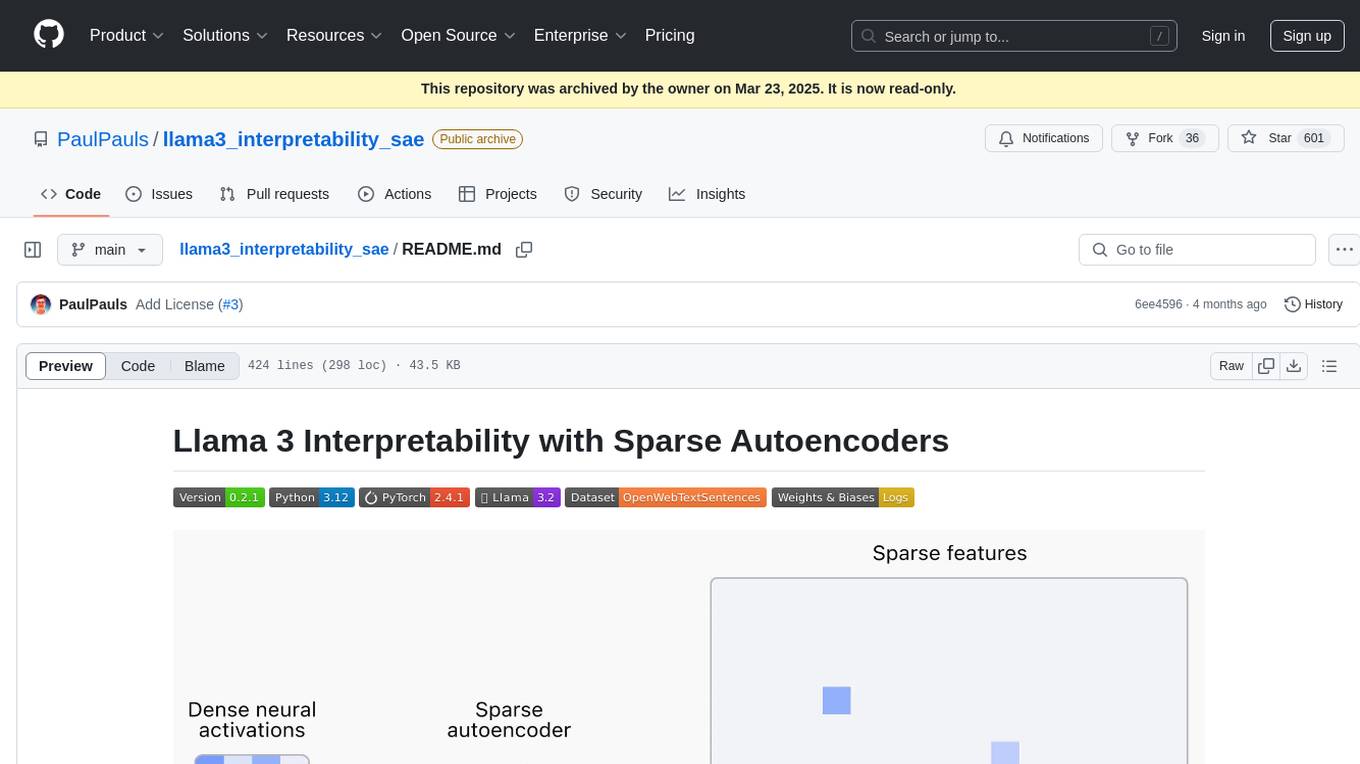
llama3_interpretability_sae
This project focuses on implementing Sparse Autoencoders (SAEs) for mechanistic interpretability in Large Language Models (LLMs) like Llama 3.2-3B. The SAEs aim to untangle superimposed representations in LLMs into separate, interpretable features for each neuron activation. The project provides an end-to-end pipeline for capturing training data, training the SAEs, analyzing learned features, and verifying results experimentally. It includes comprehensive logging, visualization, and checkpointing of SAE training, interpretability analysis tools, and a pure PyTorch implementation of Llama 3.1/3.2 chat and text completion. The project is designed for scalability, efficiency, and maintainability.
LLMSpeculativeSampling
This repository implements speculative sampling for large language model (LLM) decoding, utilizing two models - a target model and an approximation model. The approximation model generates token guesses, corrected by the target model, resulting in improved efficiency. It includes implementations of Google's and Deepmind's versions of speculative sampling, supporting models like llama-7B and llama-1B. The tool is designed for fast inference from transformers via speculative decoding.
ai-algorithms
This repository is a work in progress that contains first-principle implementations of groundbreaking AI algorithms using various deep learning frameworks. Each implementation is accompanied by supporting research papers, aiming to provide comprehensive educational resources for understanding and implementing foundational AI algorithms from scratch.
For similar tasks

LLM-for-genomics-training
This repository provides training on large language models (LLMs) for genomics, including lecture notes and lab classes covering pretraining, finetuning, zeroshot learning prediction of mutation effect, synthetic DNA sequence generation, and DNA sequence optimization.
For similar jobs

NoLabs
NoLabs is an open-source biolab that provides easy access to state-of-the-art models for bio research. It supports various tasks, including drug discovery, protein analysis, and small molecule design. NoLabs aims to accelerate bio research by making inference models accessible to everyone.

OpenCRISPR
OpenCRISPR is a set of free and open gene editing systems designed by Profluent Bio. The OpenCRISPR-1 protein maintains the prototypical architecture of a Type II Cas9 nuclease but is hundreds of mutations away from SpCas9 or any other known natural CRISPR-associated protein. You can view OpenCRISPR-1 as a drop-in replacement for many protocols that need a cas9-like protein with an NGG PAM and you can even use it with canonical SpCas9 gRNAs. OpenCRISPR-1 can be fused in a deactivated or nickase format for next generation gene editing techniques like base, prime, or epigenome editing.
ersilia
The Ersilia Model Hub is a unified platform of pre-trained AI/ML models dedicated to infectious and neglected disease research. It offers an open-source, low-code solution that provides seamless access to AI/ML models for drug discovery. Models housed in the hub come from two sources: published models from literature (with due third-party acknowledgment) and custom models developed by the Ersilia team or contributors.
ontogpt
OntoGPT is a Python package for extracting structured information from text using large language models, instruction prompts, and ontology-based grounding. It provides a command line interface and a minimal web app for easy usage. The tool has been evaluated on test data and is used in related projects like TALISMAN for gene set analysis. OntoGPT enables users to extract information from text by specifying relevant terms and provides the extracted objects as output.
bia-bob
BIA `bob` is a Jupyter-based assistant for interacting with data using large language models to generate Python code. It can utilize OpenAI's chatGPT, Google's Gemini, Helmholtz' blablador, and Ollama. Users need respective accounts to access these services. Bob can assist in code generation, bug fixing, code documentation, GPU-acceleration, and offers a no-code custom Jupyter Kernel. It provides example notebooks for various tasks like bio-image analysis, model selection, and bug fixing. Installation is recommended via conda/mamba environment. Custom endpoints like blablador and ollama can be used. Google Cloud AI API integration is also supported. The tool is extensible for Python libraries to enhance Bob's functionality.

Scientific-LLM-Survey
Scientific Large Language Models (Sci-LLMs) is a repository that collects papers on scientific large language models, focusing on biology and chemistry domains. It includes textual, molecular, protein, and genomic languages, as well as multimodal language. The repository covers various large language models for tasks such as molecule property prediction, interaction prediction, protein sequence representation, protein sequence generation/design, DNA-protein interaction prediction, and RNA prediction. It also provides datasets and benchmarks for evaluating these models. The repository aims to facilitate research and development in the field of scientific language modeling.
polaris
Polaris establishes a novel, industry‑certified standard to foster the development of impactful methods in AI-based drug discovery. This library is a Python client to interact with the Polaris Hub. It allows you to download Polaris datasets and benchmarks, evaluate a custom method against a Polaris benchmark, and create and upload new datasets and benchmarks.
awesome-AI4MolConformation-MD
The 'awesome-AI4MolConformation-MD' repository focuses on protein conformations and molecular dynamics using generative artificial intelligence and deep learning. It provides resources, reviews, datasets, packages, and tools related to AI-driven molecular dynamics simulations. The repository covers a wide range of topics such as neural networks potentials, force fields, AI engines/frameworks, trajectory analysis, visualization tools, and various AI-based models for protein conformational sampling. It serves as a comprehensive guide for researchers and practitioners interested in leveraging AI for studying molecular structures and dynamics.