
bia-bob
BIA Bob is a Jupyter+LLM-based assistant for interacting with image data and for working on Bio-image Analysis tasks.
Stars: 110
BIA `bob` is a Jupyter-based assistant for interacting with data using large language models to generate Python code. It can utilize OpenAI's chatGPT, Google's Gemini, Helmholtz' blablador, and Ollama. Users need respective accounts to access these services. Bob can assist in code generation, bug fixing, code documentation, GPU-acceleration, and offers a no-code custom Jupyter Kernel. It provides example notebooks for various tasks like bio-image analysis, model selection, and bug fixing. Installation is recommended via conda/mamba environment. Custom endpoints like blablador and ollama can be used. Google Cloud AI API integration is also supported. The tool is extensible for Python libraries to enhance Bob's functionality.
README:
BIA bob is a Jupyter-based assistant for interacting with data using large language models which generate Python code for Bio-Image Analysis (BIA).
It can make use of OpenAI's chatGPT, Google's Gemini, Anthropic's Claude, Github Models Marketplace, Helmholtz' blablador and Ollama.
You need an OpenAI API account or a Google Cloud account or a Helmholtz ID account to use it.
Using it with Ollama is free but requires running an Ollama server locally.
bob can write short Python code snippets and entire Jupyter notebooks for your image / data analysis workflow.
[!CAUTION] When using the OpenAI, Google Gemini, Anthropic, Github Models or any other endpoint via BiA-Bob, you are bound to the terms of service of the respective companies or organizations. The prompts you enter are transferred to their servers and may be processed and stored there. Make sure to not submit any sensitive, confidential or personal data. Also using these services may cost money.
You can initialize bob like this:
from bia_bob import bob
You can ask Bob to generate code like this:
%bob Load blobs.tif and show it
It will then respond with a Python code snippet that you can execute (see full example):
Use %bob if you want to write in the same line and %%bob if you want to write below.
If you want to continue using a variable in the next code cell, you need to specify the name of the variable in the following prompt.
When asking Bob explicitly to generate a notebook, it will put a new notebook file in the current directory with the generated code (See full example). You can then open it in Jupyter lab.
You can also ask Bob to modify an existing notebook, e.g. to introduce explanatory markdown cells (See full example):
Furthermore, one can translate Jupyter notebooks to other languages, e.g. by prompting %bob translate the filename.ipynb to <language>.
You can also ask Bob to write a prompt for you. This can be useful to explore potential strategies for analyzing image data. Note: It might be necessary to modify those prompts, especially when suggested analysis workflows are long and complicated. Shorten suggested prompts to the minimal necessary steps to answer your scientific question. (See full example).
You can add additional information from given Python variables into your prompt using the {variable_name} syntax.
With this, the content of the variable will become part of the prompt (full example).
If you are not sure what generated code does, you can ask Bob to explain it to you:
Bob can fix simple bugs in code you executed. Just add %%fix on top of the cell right after the error happened.
Using the %%doc magic, you can generate documentation for a given code cell.
Using the %%acc magic, you can replace common image processing functions with GPU-accelerated functions. It is recommended to check if the image processing results remain the same. You can see an example in this notebook.
You can use bia-bob from the terminal. This is recommended for creating notebooks for example like this:
bia-bob Please create a Jupyter Notebook that opens blobs.tif, segments the bright objects and shows the resulting label image on top of the original image with a curtain.
This can also be used to create other files, e.g. CSV files.
bia-bob is a research project aiming at streamlining the design of image analysis workflows. Under the hood it uses
artificial intelligence / large language models to generate text and code fulfilling the user's requests.
Users are responsible to verify the generated code according to good scientific practice. Some general advice:
- If you do not understand what a generated code snippet does, ask
%%bob explain this code in detail to a Python beginner:before executing the code. - After Bob generated a data analysis workflow for you, ask
%%bob How could I verify this analysis workflow ?. It is good scientific practice to measure the quality of segmentation results for example, or to measure the difference of automated quantitative measurements, in comparison to manual analysis. - If you are not sure if an image analysis workflow is valid, consider asking human experts. E.g. reach out via https://image.sc .
When sending a request to the LLM service providers, bia-bob sends the following information:
- The content of the cell you were typing
- If you entered variables using the
{variable}syntax, the content of these variables - All available variable, function and module names in the current Python environment (not variable values)
- A selected list of Python libraries that are installed
- The conversation history of the current session
- Optional: The image mentioned in the first line of the cell
This information is necessary to enable bia-bob to generate code that runs in your environment. If you want to know exactly what is sent to the server, you can activate verbose mode like this:
from bia_bob._machinery import Context
Context.verbose = True
If you want to ask bob a question, you need to put a space before the ?.
%bob What do you know about blobs.tif ?
You can install bia-bob using conda/pip. It is recommended to install it into a conda/mamba environment. If you have never used conda before, please read this guide first.
It is recommended to install bia-bob in a conda-environment together with useful tools for bio-image analysis.
conda env create -f https://github.com/haesleinhuepf/bia-bob/raw/main/environment.yml
You can then activate this environment...
conda activate bob_env
OR install bob into an existing environment:
pip install bia-bob
If you would like to use the %bob magic in notebooks without need for importing bia_bob first,
you can add a bia_bob.ipy file to your Jupyter startup directory ~/.ipython/profile_default/startup/ with this content:
from bia_bob import bob
Note: This might require to install bia_bob in the base environment.
For using LLMs from remote service providers, you need to set an API key.
API Keys are short cryptic texts such as "proj_sk_asdasdasd" which allow you to log into a remote service without entering your username and password. Many online serives require using API keys for billing; others enable you to use their free services only after obtaining an API key.
This also means that you should not share your API key with others.
In the following sections, you find links to a couple of LLM services providers that are compatible with bia-bob.
After obtaining the key, you need to add it to the enviroment variables of your computer.
On Windows, you can do this by 1) searching for "env" in the start menu, 2) clicking on "Edit the system environment variables",
3) clicking on "Environment Variables", 4) clicking on "New" in the "System variables" section and adding a new variable with the name specified below (e.g. OPENAI_API_KEY) and the value of your API key.
On Linux and MacOS, this is typically done by modifying a hidden .bashrc or .zshrc file in the home directory, e.g. like this:
echo "export OPENAI_API_KEY='yourkey'" >> ~/.zshrc
Note: After setting the environment variables, you need to restart your terminal and/or Jupyter Lab to make them work.
See also further instructions on this page.
Create an OpenAI API Key and add it to your environment variables named OPENAI_API_KEY as explained on
You can then initialize Bob like this (optional, as that's default):
from bia_bob import bob
bob.initialize("gpt-4o-2024-08-06", vision_model="gpt-4o-2024-08-06")
Create an Anthropic API Key and add it to your environment variables named ANTHROPIC_API_KEY.
You can then initialize Bob like this:
from bia_bob import bob
bob.initialize(model="claude-3-5-sonnet-20240620", vision_model="claude-3-5-sonnet-20240620")
You can also apply for an API Key from the German Artificial Intelligence Service Center for Sensible and Critical Infrastructures who operates the ChatAI service.
You can store it in an environment variable named OPENAI_API_KEY and use initialize bob like this:
from bia_bob import bob
bob.initialize(endpoint="https://chat-ai.academiccloud.de/v1",
model="meta-llama-3.1-70b-instruct")
If you are using the models from Github Models Marketplace, please create an GITHUB API key (with default settings) and store it for accessing the models in an environment variable named GH_MODELS_API_KEY.
You can then access the models like this:
bob.initialize(
endpoint='github_models',
model='Phi-3.5-mini-instruct')
If you are using the models hosted on Microsoft Azure, please store your API key for accessing the models in an environment variable named AZURE_API_KEY.
You can then access the models like this:
bob.initialize(
endpoint='azure',
model='Phi-3.5-mini-instruct')
Alternatively, you can specify the endpoint directly, too:
bob.initialize(
endpoint='https://models.inference.ai.azure.com',
model='Phi-3.5-mini-instruct')
Custom endpoints can be used as well if they support the OpenAI API. Examples are DeepSeek, KISSKI, blablador and ollama. An example is shown in this notebook:
For this, just install the openai backend as explained above (tested version: 1.5.0).
- If you want to use ollama and e.g. the
codellamamodel, you must runollama servefrom a separate terminal and then initialize bob like this:
bob.initialize(endpoint='ollama', model='codellama')
- For using DeepSeek, you need to get an API key. Store it in your environment as
DEEPSEEK_API_KEYvariable.
bob.initialize(endpoint='deepseek', model='deepseek-chat')
- If you want to use blablador, which is free for German academics, just get an API key as explained on
this page and store it in your environment as
BLABLADOR_API_KEYvariable.
bob.initialize(
endpoint='blablador',
model='Mistral-7B-Instruct-v0.2')
- Custom end points can be used as well, for example like this:
bob.initialize(
endpoint='http://localhost:11434/v1',
model='codellama')
Create a Google API key and store it in the environment variable GOOGLE_API_KEY.
pip install google-generativeai>=0.7.2
You can then initialize Bob like this:
from bia_bob import bob
bob.initialize("gemini-1.5-pro-002")
Note: This method is deprecated. Use gemini 1.5 as shown above.
pip install google-cloud-aiplatform
(Recommended google-cloud-aiplatform version >= 1.38.1)
To make use of the Google Cloud API, you need to create a Google Cloud account here and a project within the Google cloud (for billing) here. You need to store authentication details locally as explained here. This requires installing Google Cloud CLI. In very short: run the installer and when asked, activate the "Run gcloud init" checkbox. Or run 'gcloud init' from the terminal yourself. Restart the terminal window. After installing Google Cloud CLI, start a terminal and authenticate using:
gcloud auth application-default login
Follow the instructions in the browser. Enter your Project ID (not the name). If it worked the terminal should approximately look like this:
You can also configure the system message bia-bob is using, which might be useful in particular when using it with
small open-weight language models executed on local hardware, or when using it in scenarios that are not bio-image
analysis related.
from bia_bob import bob
bob.initialize(system_prompt="""
You are an excellent astronomer and Python programmer.
You typically use Python libraris from this domain.
""")
In this system message you can use place-holders which are set when bob is invoked:
-
{libraries}: A list of Python libraries that are installed an bio-image analysis related (scikit-image, stackview, ...) -
{reusable_variables}: A list of variables that are available in the current Python environment -
{builtin_snippets}: A list of built-in code snippets that might be helpful for bio-image analysis -
{additional_snippets}: A list of additional code snippets that is assembled from Python libraries that support giving hints to bob.
More examples are available in the demo notebook.
If you want to contribute to bia-bob, you can install it in development mode like this:
git clone https://github.com/haesleinhuepf/bia-bob.git
cd bia-bob
pip install -e .
If you are maintainer of a Python library and want to make BiA-bob aware of functions in your library, you can extend Bob's knowledge using entry-points. Add this to your library setup.cfg:
[options.entry_points]
bia_bob_plugins =
plugin1 = your_library._bia_bob_plugins:list_bia_bob_plugins
In the above mentioned _bia_bob_plugins.py define this function (and feel free to rename the function and the Python file):
def list_bia_bob_plugins():
"""List of function hints for bia_bob"""
return """
* Computes the sum of a and b
your_library.compute_sum(a:int,b:int) -> int
* Determines the difference between a and b
your_library.compute_difference(a:int, b:int) -> int
"""
Note that the syntax should be pretty much as shown above: A bullet point with a short description and a code-snippet just below.
You can also generate the list_bia_bob_plugins function as demonstrated in this notebook.
Please only list the most important functions. If the list of all plugins extending BiA-Bob becomes too long, the prompt will exceed the maximum prompt length.
List of known Python libraries that provide extensions to Bob:
(Feel free to extend this list by sending a pull-request)
There are similar projects:
- jupyter-ai
- JupyterLab Magic Wand
- chatGPT-jupyter-extension
- chapyter
- napari-chatGPT
- bioimageio-chatbot
- Claude Engineer
- BioChatter
- aider
- OpenDevin
- Devika
If you encounter any problems or want to provide feedback or suggestions, please create a thread on image.sc along with a detailed description and tag @haesleinhuepf .
For Tasks:
Click tags to check more tools for each tasksFor Jobs:
Alternative AI tools for bia-bob
Similar Open Source Tools

bia-bob
BIA `bob` is a Jupyter-based assistant for interacting with data using large language models to generate Python code. It can utilize OpenAI's chatGPT, Google's Gemini, Helmholtz' blablador, and Ollama. Users need respective accounts to access these services. Bob can assist in code generation, bug fixing, code documentation, GPU-acceleration, and offers a no-code custom Jupyter Kernel. It provides example notebooks for various tasks like bio-image analysis, model selection, and bug fixing. Installation is recommended via conda/mamba environment. Custom endpoints like blablador and ollama can be used. Google Cloud AI API integration is also supported. The tool is extensible for Python libraries to enhance Bob's functionality.
torchchat
torchchat is a codebase showcasing the ability to run large language models (LLMs) seamlessly. It allows running LLMs using Python in various environments such as desktop, server, iOS, and Android. The tool supports running models via PyTorch, chatting, generating text, running chat in the browser, and running models on desktop/server without Python. It also provides features like AOT Inductor for faster execution, running in C++ using the runner, and deploying and running on iOS and Android. The tool supports popular hardware and OS including Linux, Mac OS, Android, and iOS, with various data types and execution modes available.
sage
Sage is a tool that allows users to chat with any codebase, providing a chat interface for code understanding and integration. It simplifies the process of learning how a codebase works by offering heavily documented answers sourced directly from the code. Users can set up Sage locally or on the cloud with minimal effort. The tool is designed to be easily customizable, allowing users to swap components of the pipeline and improve the algorithms powering code understanding and generation.
WindowsAgentArena
Windows Agent Arena (WAA) is a scalable Windows AI agent platform designed for testing and benchmarking multi-modal, desktop AI agents. It provides researchers and developers with a reproducible and realistic Windows OS environment for AI research, enabling testing of agentic AI workflows across various tasks. WAA supports deploying agents at scale using Azure ML cloud infrastructure, allowing parallel running of multiple agents and delivering quick benchmark results for hundreds of tasks in minutes.
fabric
Fabric is an open-source framework for augmenting humans using AI. It provides a structured approach to breaking down problems into individual components and applying AI to them one at a time. Fabric includes a collection of pre-defined Patterns (prompts) that can be used for a variety of tasks, such as extracting the most interesting parts of YouTube videos and podcasts, writing essays, summarizing academic papers, creating AI art prompts, and more. Users can also create their own custom Patterns. Fabric is designed to be easy to use, with a command-line interface and a variety of helper apps. It is also extensible, allowing users to integrate it with their own AI applications and infrastructure.

fasttrackml
FastTrackML is an experiment tracking server focused on speed and scalability, fully compatible with MLFlow. It provides a user-friendly interface to track and visualize your machine learning experiments, making it easy to compare different models and identify the best performing ones. FastTrackML is open source and can be easily installed and run with pip or Docker. It is also compatible with the MLFlow Python package, making it easy to integrate with your existing MLFlow workflows.
ray-llm
RayLLM (formerly known as Aviary) is an LLM serving solution that makes it easy to deploy and manage a variety of open source LLMs, built on Ray Serve. It provides an extensive suite of pre-configured open source LLMs, with defaults that work out of the box. RayLLM supports Transformer models hosted on Hugging Face Hub or present on local disk. It simplifies the deployment of multiple LLMs, the addition of new LLMs, and offers unique autoscaling support, including scale-to-zero. RayLLM fully supports multi-GPU & multi-node model deployments and offers high performance features like continuous batching, quantization and streaming. It provides a REST API that is similar to OpenAI's to make it easy to migrate and cross test them. RayLLM supports multiple LLM backends out of the box, including vLLM and TensorRT-LLM.
vscode-pddl
The vscode-pddl extension provides comprehensive support for Planning Domain Description Language (PDDL) in Visual Studio Code. It enables users to model planning domains, validate them, industrialize planning solutions, and run planners. The extension offers features like syntax highlighting, auto-completion, plan visualization, plan validation, plan happenings evaluation, search debugging, and integration with Planning.Domains. Users can create PDDL files, run planners, visualize plans, and debug search algorithms efficiently within VS Code.
vectara-answer
Vectara Answer is a sample app for Vectara-powered Summarized Semantic Search (or question-answering) with advanced configuration options. For examples of what you can build with Vectara Answer, check out Ask News, LegalAid, or any of the other demo applications.
Bard-API
The Bard API is a Python package that returns responses from Google Bard through the value of a cookie. It is an unofficial API that operates through reverse-engineering, utilizing cookie values to interact with Google Bard for users struggling with frequent authentication problems or unable to authenticate via Google Authentication. The Bard API is not a free service, but rather a tool provided to assist developers with testing certain functionalities due to the delayed development and release of Google Bard's API. It has been designed with a lightweight structure that can easily adapt to the emergence of an official API. Therefore, using it for any other purposes is strongly discouraged. If you have access to a reliable official PaLM-2 API or Google Generative AI API, replace the provided response with the corresponding official code. Check out https://github.com/dsdanielpark/Bard-API/issues/262.
smartcat
Smartcat is a CLI interface that brings language models into the Unix ecosystem, allowing power users to leverage the capabilities of LLMs in their daily workflows. It features a minimalist design, seamless integration with terminal and editor workflows, and customizable prompts for specific tasks. Smartcat currently supports OpenAI, Mistral AI, and Anthropic APIs, providing access to a range of language models. With its ability to manipulate file and text streams, integrate with editors, and offer configurable settings, Smartcat empowers users to automate tasks, enhance code quality, and explore creative possibilities.
llamafile
llamafile is a tool that enables users to distribute and run Large Language Models (LLMs) with a single file. It combines llama.cpp with Cosmopolitan Libc to create a framework that simplifies the complexity of LLMs into a single-file executable called a 'llamafile'. Users can run these executable files locally on most computers without the need for installation, making open LLMs more accessible to developers and end users. llamafile also provides example llamafiles for various LLM models, allowing users to try out different LLMs locally. The tool supports multiple CPU microarchitectures, CPU architectures, and operating systems, making it versatile and easy to use.
HuggingFaceGuidedTourForMac
HuggingFaceGuidedTourForMac is a guided tour on how to install optimized pytorch and optionally Apple's new MLX, JAX, and TensorFlow on Apple Silicon Macs. The repository provides steps to install homebrew, pytorch with MPS support, MLX, JAX, TensorFlow, and Jupyter lab. It also includes instructions on running large language models using HuggingFace transformers. The repository aims to help users set up their Macs for deep learning experiments with optimized performance.
superflows
Superflows is an open-source alternative to OpenAI's Assistant API. It allows developers to easily add an AI assistant to their software products, enabling users to ask questions in natural language and receive answers or have tasks completed by making API calls. Superflows can analyze data, create plots, answer questions based on static knowledge, and even write code. It features a developer dashboard for configuration and testing, stateful streaming API, UI components, and support for multiple LLMs. Superflows can be set up in the cloud or self-hosted, and it provides comprehensive documentation and support.

verifAI
VerifAI is a document-based question-answering system that addresses hallucinations in generative large language models and search engines. It retrieves relevant documents, generates answers with references, and verifies answers for accuracy. The engine uses generative search technology and a verification model to ensure no misinformation. VerifAI supports various document formats and offers user registration with a React.js interface. It is open-source and designed to be user-friendly, making it accessible for anyone to use.
linkedin-api
The Linkedin API for Python allows users to programmatically search profiles, send messages, and find jobs using a regular Linkedin user account. It does not require 'official' API access, just a valid Linkedin account. However, it is important to note that this library is not officially supported by LinkedIn and using it may violate LinkedIn's Terms of Service. Users can authenticate using any Linkedin account credentials and access features like getting profiles, profile contact info, and connections. The library also provides commercial alternatives for extracting data, scraping public profiles, and accessing a full LinkedIn API. It is not endorsed or supported by LinkedIn and is intended for educational purposes and personal use only.
For similar tasks

bia-bob
BIA `bob` is a Jupyter-based assistant for interacting with data using large language models to generate Python code. It can utilize OpenAI's chatGPT, Google's Gemini, Helmholtz' blablador, and Ollama. Users need respective accounts to access these services. Bob can assist in code generation, bug fixing, code documentation, GPU-acceleration, and offers a no-code custom Jupyter Kernel. It provides example notebooks for various tasks like bio-image analysis, model selection, and bug fixing. Installation is recommended via conda/mamba environment. Custom endpoints like blablador and ollama can be used. Google Cloud AI API integration is also supported. The tool is extensible for Python libraries to enhance Bob's functionality.
cameratrapai
SpeciesNet is an ensemble of AI models designed for classifying wildlife in camera trap images. It consists of an object detector that finds objects of interest in wildlife camera images and an image classifier that classifies those objects to the species level. The ensemble combines these two models using heuristics and geographic information to assign each image to a single category. The models have been trained on a large dataset of camera trap images and are used for species recognition in the Wildlife Insights platform.
cody
Cody is a free, open-source AI coding assistant that can write and fix code, provide AI-generated autocomplete, and answer your coding questions. Cody fetches relevant code context from across your entire codebase to write better code that uses more of your codebase's APIs, impls, and idioms, with less hallucination.
auto-dev-vscode
AutoDev for VSCode is an AI-powered coding wizard with multilingual support, auto code generation, and a bug-slaying assistant. It offers customizable prompts and features like Auto Dev/Testing/Document/Agent. The tool aims to enhance coding productivity and efficiency by providing intelligent assistance and automation capabilities within the Visual Studio Code environment.
code2prompt
code2prompt is a command-line tool that converts your codebase into a single LLM prompt with a source tree, prompt templating, and token counting. It automates generating LLM prompts from codebases of any size, customizing prompt generation with Handlebars templates, respecting .gitignore, filtering and excluding files using glob patterns, displaying token count, including Git diff output, copying prompt to clipboard, saving prompt to an output file, excluding files and folders, adding line numbers to source code blocks, and more. It helps streamline the process of creating LLM prompts for code analysis, generation, and other tasks.
fittencode.nvim
Fitten Code AI Programming Assistant for Neovim provides fast completion using AI, asynchronous I/O, and support for various actions like document code, edit code, explain code, find bugs, generate unit test, implement features, optimize code, refactor code, start chat, and more. It offers features like accepting suggestions with Tab, accepting line with Ctrl + Down, accepting word with Ctrl + Right, undoing accepted text, automatic scrolling, and multiple HTTP/REST backends. It can run as a coc.nvim source or nvim-cmp source.
chatgpt
The ChatGPT R package provides a set of features to assist in R coding. It includes addins like Ask ChatGPT, Comment selected code, Complete selected code, Create unit tests, Create variable name, Document code, Explain selected code, Find issues in the selected code, Optimize selected code, and Refactor selected code. Users can interact with ChatGPT to get code suggestions, explanations, and optimizations. The package helps in improving coding efficiency and quality by providing AI-powered assistance within the RStudio environment.
VimLM
VimLM is an AI-powered coding assistant for Vim that integrates AI for code generation, refactoring, and documentation directly into your Vim workflow. It offers native Vim integration with split-window responses and intuitive keybindings, offline first execution with MLX-compatible models, contextual awareness with seamless integration with codebase and external resources, conversational workflow for iterating on responses, project scaffolding for generating and deploying code blocks, and extensibility for creating custom LLM workflows with command chains.
For similar jobs

NoLabs
NoLabs is an open-source biolab that provides easy access to state-of-the-art models for bio research. It supports various tasks, including drug discovery, protein analysis, and small molecule design. NoLabs aims to accelerate bio research by making inference models accessible to everyone.

OpenCRISPR
OpenCRISPR is a set of free and open gene editing systems designed by Profluent Bio. The OpenCRISPR-1 protein maintains the prototypical architecture of a Type II Cas9 nuclease but is hundreds of mutations away from SpCas9 or any other known natural CRISPR-associated protein. You can view OpenCRISPR-1 as a drop-in replacement for many protocols that need a cas9-like protein with an NGG PAM and you can even use it with canonical SpCas9 gRNAs. OpenCRISPR-1 can be fused in a deactivated or nickase format for next generation gene editing techniques like base, prime, or epigenome editing.
ersilia
The Ersilia Model Hub is a unified platform of pre-trained AI/ML models dedicated to infectious and neglected disease research. It offers an open-source, low-code solution that provides seamless access to AI/ML models for drug discovery. Models housed in the hub come from two sources: published models from literature (with due third-party acknowledgment) and custom models developed by the Ersilia team or contributors.
ontogpt
OntoGPT is a Python package for extracting structured information from text using large language models, instruction prompts, and ontology-based grounding. It provides a command line interface and a minimal web app for easy usage. The tool has been evaluated on test data and is used in related projects like TALISMAN for gene set analysis. OntoGPT enables users to extract information from text by specifying relevant terms and provides the extracted objects as output.

bia-bob
BIA `bob` is a Jupyter-based assistant for interacting with data using large language models to generate Python code. It can utilize OpenAI's chatGPT, Google's Gemini, Helmholtz' blablador, and Ollama. Users need respective accounts to access these services. Bob can assist in code generation, bug fixing, code documentation, GPU-acceleration, and offers a no-code custom Jupyter Kernel. It provides example notebooks for various tasks like bio-image analysis, model selection, and bug fixing. Installation is recommended via conda/mamba environment. Custom endpoints like blablador and ollama can be used. Google Cloud AI API integration is also supported. The tool is extensible for Python libraries to enhance Bob's functionality.

Scientific-LLM-Survey
Scientific Large Language Models (Sci-LLMs) is a repository that collects papers on scientific large language models, focusing on biology and chemistry domains. It includes textual, molecular, protein, and genomic languages, as well as multimodal language. The repository covers various large language models for tasks such as molecule property prediction, interaction prediction, protein sequence representation, protein sequence generation/design, DNA-protein interaction prediction, and RNA prediction. It also provides datasets and benchmarks for evaluating these models. The repository aims to facilitate research and development in the field of scientific language modeling.
polaris
Polaris establishes a novel, industry‑certified standard to foster the development of impactful methods in AI-based drug discovery. This library is a Python client to interact with the Polaris Hub. It allows you to download Polaris datasets and benchmarks, evaluate a custom method against a Polaris benchmark, and create and upload new datasets and benchmarks.
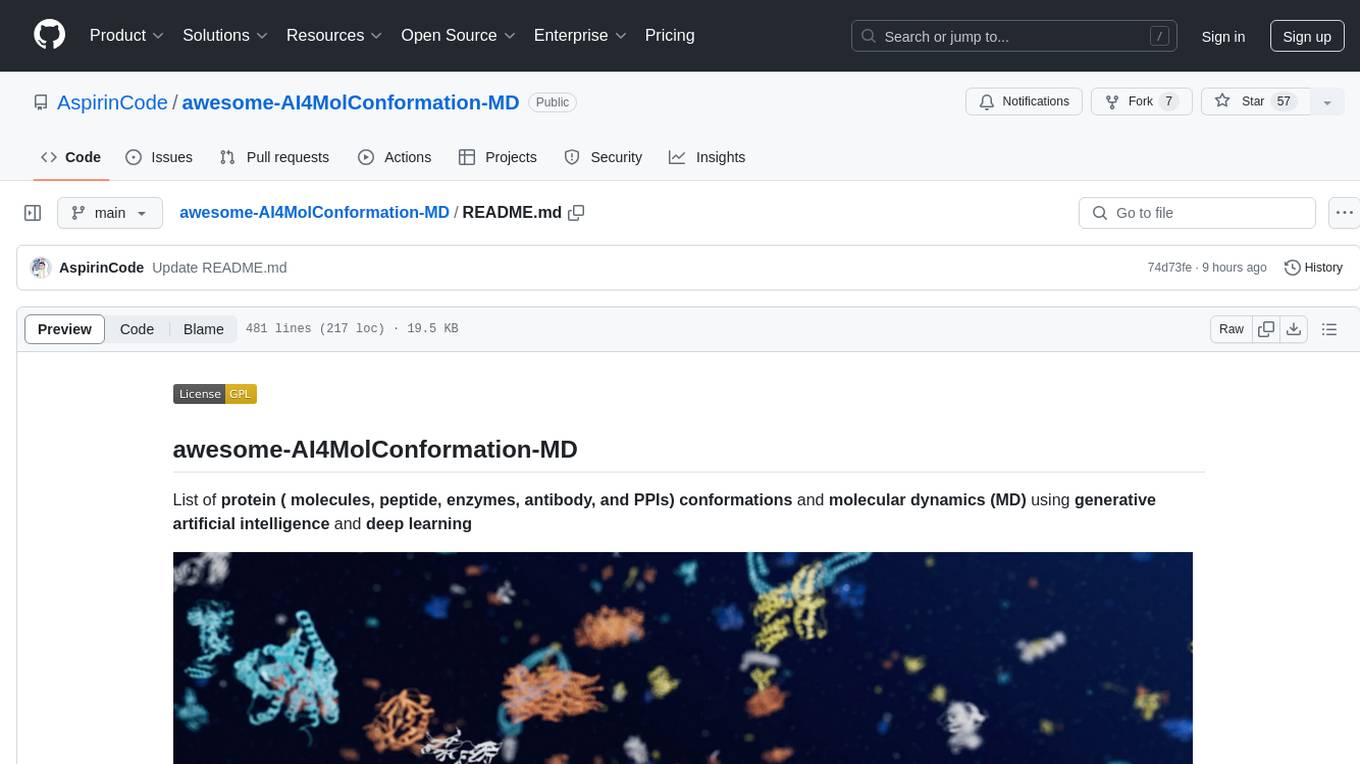
awesome-AI4MolConformation-MD
The 'awesome-AI4MolConformation-MD' repository focuses on protein conformations and molecular dynamics using generative artificial intelligence and deep learning. It provides resources, reviews, datasets, packages, and tools related to AI-driven molecular dynamics simulations. The repository covers a wide range of topics such as neural networks potentials, force fields, AI engines/frameworks, trajectory analysis, visualization tools, and various AI-based models for protein conformational sampling. It serves as a comprehensive guide for researchers and practitioners interested in leveraging AI for studying molecular structures and dynamics.





















