
rtdl-num-embeddings
(NeurIPS 2022) On Embeddings for Numerical Features in Tabular Deep Learning
Stars: 287
This repository provides the official implementation of the paper 'On Embeddings for Numerical Features in Tabular Deep Learning'. It focuses on transforming scalar continuous features into vectors before integrating them into the main backbone of tabular neural networks, showcasing improved performance. The embeddings for continuous features are shown to enhance the performance of tabular DL models and are applicable to various conventional backbones, offering efficiency comparable to Transformer-based models. The repository includes Python packages for practical usage, exploration of metrics and hyperparameters, and reproducing reported results for different algorithms and datasets.
README:
📜 arXiv 📦 Python package 📚 RTDL (other projects on tabular DL)
This is the official implementation of the paper "On Embeddings for Numerical Features in Tabular Deep Learning".
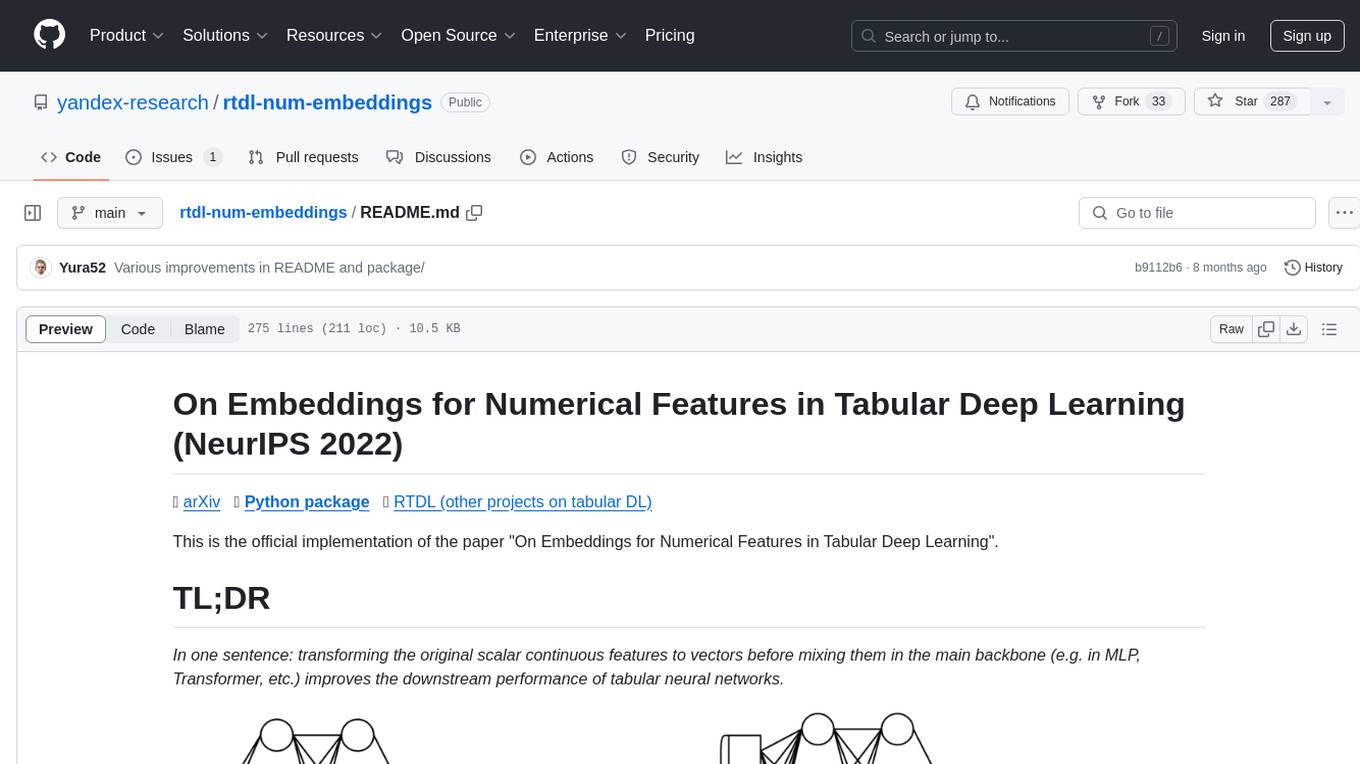
In one sentence: transforming the original scalar continuous features to vectors before mixing them in the main backbone (e.g. in MLP, Transformer, etc.) improves the downstream performance of tabular neural networks.
Left: vanilla MLP taking two continuous features as input.
Right: the same MLP, but now with embeddings for continuous features.
In more detail:
- Embedding continuous features means transforming them from scalar representations to vectors before mixing in the main backbone as illustrated above.
- It turns out that embeddings for continuous features can (significantly) improve the performance of tabular DL models.
- Embeddings are applicable to any conventional backbone.
- In particular, simple MLP with embeddings can be competitive with heavy Transormer-based models while being significantly more efficient.
- Despite the formal overhead in terms of parameter count, in practice, embeddings are perfectly affordable in many cases. On big enough datasets and/or with large enough number of features and/or with strict enough latency requirements, the new overhead associated with embeddings may become an issue.
Why do embeddings work?
Strictly speaking, there is no single explanation. Evidently, the embeddings help dealing with various challenges associated with continuous features and improve the overall optimization properties of models.
In particular, irregularly distributed continuous features (and their irregular joint distributions with labels) is a usual thing in real world tabular data, and they pose a major fundamental optimization challenge for traditional tabular DL models. A great reference for understanding this challenge (and a great example of addressing those challenges by transforming input space) is the paper "Fourier Features Let Networks Learn High Frequency Functions in Low Dimensional Domains".
However, it is unclear whether irregular distributions is the only reason why the embeddings are useful.
The Python package in the package/ directory
is the recommended way to use the paper in practice and for future work.
The rest of the document:
The exp/ directory contains numerious results and (tuned) hyperparameters
for various models and datasets used in the paper.
For example, let's explore the metrics for the MLP model.
First, let's load the reports (the report.json files):
import json
from pathlib import Path
import pandas as pd
df = pd.json_normalize([
json.loads(x.read_text())
for x in Path('exp').glob('mlp/*/0_evaluation/*/report.json')
])Now, for each dataset, let's compute the test score averaged over all random seeds:
print(df.groupby('config.data.path')['metrics.test.score'].mean().round(3))The output exactly matches Table 3 from the paper:
config.data.path
data/adult 0.854
data/california -0.495
data/churn 0.856
data/covtype 0.964
data/fb-comments -5.686
data/gesture 0.632
data/higgs-small 0.720
data/house -32039.399
data/microsoft -0.747
data/otto 0.818
data/santander 0.912
Name: metrics.test.score, dtype: float64
The above approach can also be used to explore hyperparameters to get intuition on typical hyperparameter values for different algorithms. For example, this is how one can compute the median tuned learning rate for the MLP model:
[!NOTE] For some algorithms (e.g. MLP, MLP-LR, MLP-PLR), more recent projects offer more results that can be explored in a similar way. For example, see this paper on TabR.
[!WARNING] Use this approach with caution. When studying hyperparameter values:
- Beware of outliers.
- Take a look at raw unaggregated values to get intuition on typical values.
- For a high-level overview, plot the distribution and/or compute multiple quantiles.
print(df[df['config.seed'] == 0]['config.training.lr'].quantile(0.5))
# Output: 0.0002716544410603358[!IMPORTANT]
This section is long. Use the "Outline" feature on GitHub on in your text editor to get an overview of this section.
Preliminaries:
- You may need to change the CUDA-related commands and settings below depending on your setup
- Make sure that
/usr/local/cuda-11.1/binis always in yourPATHenvironment variable - Install conda
export PROJECT_DIR=<ABSOLUTE path to the repository root>
# example: export PROJECT_DIR=/home/myusername/repositories/num-embeddings
git clone https://github.com/yandex-research/tabular-dl-num-embeddings $PROJECT_DIR
cd $PROJECT_DIR
conda create -n num-embeddings python=3.9.7
conda activate num-embeddings
pip install torch==1.10.1+cu111 -f https://download.pytorch.org/whl/torch_stable.html
pip install -r requirements.txt
# if the following commands do not succeed, update conda
conda env config vars set PYTHONPATH=${PYTHONPATH}:${PROJECT_DIR}
conda env config vars set PROJECT_DIR=${PROJECT_DIR}
# the following command appends ":/usr/local/cuda-11.1/lib64" to LD_LIBRARY_PATH;
# if your LD_LIBRARY_PATH already contains a path to some other CUDA, then the content
# after "=" should be "<your LD_LIBRARY_PATH without your cuda path>:/usr/local/cuda-11.1/lib64"
conda env config vars set LD_LIBRARY_PATH=${LD_LIBRARY_PATH}:/usr/local/cuda-11.1/lib64
conda env config vars set CUDA_HOME=/usr/local/cuda-11.1
conda env config vars set CUDA_ROOT=/usr/local/cuda-11.1
# (optional) get a shortcut for toggling the dark mode with cmd+y
conda install nodejs
jupyter labextension install jupyterlab-theme-toggle
conda deactivate
conda activate num-embeddingsLICENSE: by downloading our dataset you accept the licenses of all its components. We do not impose any new restrictions in addition to those licenses. You can find the list of sources in the paper.
cd $PROJECT_DIR
wget "https://www.dropbox.com/s/r0ef3ij3wl049gl/data.tar?dl=1" -O num_embeddings_data.tar
tar -xvf num_embeddings_data.tarThe code below reproduces the results for MLP on the California Housing dataset. The pipeline for other algorithms and datasets is absolutely the same.
# You must explicitly set CUDA_VISIBLE_DEVICES if you want to use GPU
export CUDA_VISIBLE_DEVICES="0"
# Create a copy of the 'official' config
cp exp/mlp/california/0_tuning.toml exp/mlp/california/1_tuning.toml
# Run tuning (on GPU, it takes ~30-60min)
python bin/tune.py exp/mlp/california/1_tuning.toml
# Evaluate single models with 15 different random seeds
python bin/evaluate.py exp/mlp/california/1_tuning 15
# Evaluate ensembles (by default, three ensembles of size five each)
python bin/ensemble.py exp/mlp/california/1_evaluation
# Then use bin/results.ipynb to view the obtained results
The code is organized as follows:
-
bin-
train4.pyfor neural networks (it implements all the embeddings and backbones from the paper) -
xgboost_.pyfor XGBoost -
catboost_.pyfor CatBoost -
tune.pyfor tuning -
evaluate.pyfor evaluation -
ensemble.pyfor ensembling -
results.ipynbfor summarizing results -
datasets.pywas used to build the dataset splits -
synthetic.pyfor generating the synthetic GBDT-friendly datasets -
train1_synthetic.pyfor the experiments with synthetic data
-
-
libcontains common tools used by programs inbin -
expcontains experiment configs and results (metrics, tuned configurations, etc.). The names of the nested folders follow the names from the paper (example:exp/mlp-plrcorresponds to the MLP-PLR model from the paper). -
packagecontains the Python package for this paper
- You must explicitly set
CUDA_VISIBLE_DEVICESwhen running scripts - for saving and loading configs, use
lib.dump_configandlib.load_configinstead of bare TOML libraries
The common pattern for running scripts is:
python bin/my_script.py a/b/c.tomlwhere a/b/c.toml is the input configuration file (config). The output will be located at a/b/c. The config structure usually follows the Config class from bin/my_script.py.
There are also scripts that take command line arguments instead of configs (e.g. bin/{evaluate.py,ensemble.py}).
You need all of them for reproducing results, but you need only train4.py for future work, because:
-
bin/train1.pyimplements a superset of features frombin/train0.py -
bin/train3.pyimplements a superset of features frombin/train1.py -
bin/train4.pyimplements a superset of features frombin/train3.py
To see which one of the four scripts was used to run a given experiment, check the "program" field of the corresponding tuning config. For example, here is the tuning config for MLP on the California Housing dataset: exp/mlp/california/0_tuning.toml. The config indicates that bin/train0.py was used. It means that the configs in exp/mlp/california/0_evaluation are compatible specifically with bin/train0.py. To verify that, you can copy one of them to a separate location and pass to bin/train0.py:
mkdir exp/tmp
cp exp/mlp/california/0_evaluation/0.toml exp/tmp/0.toml
python bin/train0.py exp/tmp/0.toml
ls exp/tmp/0
@inproceedings{gorishniy2022embeddings,
title={On Embeddings for Numerical Features in Tabular Deep Learning},
author={Yury Gorishniy and Ivan Rubachev and Artem Babenko},
booktitle={{NeurIPS}},
year={2022},
}
For Tasks:
Click tags to check more tools for each tasksFor Jobs:
Alternative AI tools for rtdl-num-embeddings
Similar Open Source Tools

rtdl-num-embeddings
This repository provides the official implementation of the paper 'On Embeddings for Numerical Features in Tabular Deep Learning'. It focuses on transforming scalar continuous features into vectors before integrating them into the main backbone of tabular neural networks, showcasing improved performance. The embeddings for continuous features are shown to enhance the performance of tabular DL models and are applicable to various conventional backbones, offering efficiency comparable to Transformer-based models. The repository includes Python packages for practical usage, exploration of metrics and hyperparameters, and reproducing reported results for different algorithms and datasets.
generative-models
Generative Models by Stability AI is a repository that provides various generative models for research purposes. It includes models like Stable Video 4D (SV4D) for video synthesis, Stable Video 3D (SV3D) for multi-view synthesis, SDXL-Turbo for text-to-image generation, and more. The repository focuses on modularity and implements a config-driven approach for building and combining submodules. It supports training with PyTorch Lightning and offers inference demos for different models. Users can access pre-trained models like SDXL-base-1.0 and SDXL-refiner-1.0 under a CreativeML Open RAIL++-M license. The codebase also includes tools for invisible watermark detection in generated images.
safety-tooling
This repository, safety-tooling, is designed to be shared across various AI Safety projects. It provides an LLM API with a common interface for OpenAI, Anthropic, and Google models. The aim is to facilitate collaboration among AI Safety researchers, especially those with limited software engineering backgrounds, by offering a platform for contributing to a larger codebase. The repo can be used as a git submodule for easy collaboration and updates. It also supports pip installation for convenience. The repository includes features for installation, secrets management, linting, formatting, Redis configuration, testing, dependency management, inference, finetuning, API usage tracking, and various utilities for data processing and experimentation.

lhotse
Lhotse is a Python library designed to make speech and audio data preparation flexible and accessible. It aims to attract a wider community to speech processing tasks by providing a Python-centric design and an expressive command-line interface. Lhotse offers standard data preparation recipes, PyTorch Dataset classes for speech tasks, and efficient data preparation for model training with audio cuts. It supports data augmentation, feature extraction, and feature-space cut mixing. The tool extends Kaldi's data preparation recipes with seamless PyTorch integration, human-readable text manifests, and convenient Python classes.
probsem
ProbSem is a repository that provides a framework to leverage large language models (LLMs) for assigning context-conditional probability distributions over queried strings. It supports OpenAI engines and HuggingFace CausalLM models, and is flexible for research applications in linguistics, cognitive science, program synthesis, and NLP. Users can define prompts, contexts, and queries to derive probability distributions over possible completions, enabling tasks like cloze completion, multiple-choice QA, semantic parsing, and code completion. The repository offers CLI and API interfaces for evaluation, with options to customize models, normalize scores, and adjust temperature for probability distributions.
strwythura
Strwythura is a library and tutorial focused on constructing a knowledge graph from unstructured data sources using state-of-the-art models for named entity recognition. It implements an enhanced GraphRAG approach and curates semantics for optimizing AI application outcomes within a specific domain. The tutorial emphasizes the use of sophisticated NLP pipelines based on spaCy, GLiNER, TextRank, and related libraries to provide better/faster/cheaper results with more control over the intentional arrangement of the knowledge graph. It leverages neurosymbolic AI methods and combines practices from natural language processing, graph data science, entity resolution, ontology pipeline, context engineering, and human-in-the-loop processes.
LLMeBench
LLMeBench is a flexible framework designed for accelerating benchmarking of Large Language Models (LLMs) in the field of Natural Language Processing (NLP). It supports evaluation of various NLP tasks using model providers like OpenAI, HuggingFace Inference API, and Petals. The framework is customizable for different NLP tasks, LLM models, and datasets across multiple languages. It features extensive caching capabilities, supports zero- and few-shot learning paradigms, and allows on-the-fly dataset download and caching. LLMeBench is open-source and continuously expanding to support new models accessible through APIs.
mosec
Mosec is a high-performance and flexible model serving framework for building ML model-enabled backend and microservices. It bridges the gap between any machine learning models you just trained and the efficient online service API. * **Highly performant** : web layer and task coordination built with Rust 🦀, which offers blazing speed in addition to efficient CPU utilization powered by async I/O * **Ease of use** : user interface purely in Python 🐍, by which users can serve their models in an ML framework-agnostic manner using the same code as they do for offline testing * **Dynamic batching** : aggregate requests from different users for batched inference and distribute results back * **Pipelined stages** : spawn multiple processes for pipelined stages to handle CPU/GPU/IO mixed workloads * **Cloud friendly** : designed to run in the cloud, with the model warmup, graceful shutdown, and Prometheus monitoring metrics, easily managed by Kubernetes or any container orchestration systems * **Do one thing well** : focus on the online serving part, users can pay attention to the model optimization and business logic
kvpress
This repository implements multiple key-value cache pruning methods and benchmarks using transformers, aiming to simplify the development of new methods for researchers and developers in the field of long-context language models. It provides a set of 'presses' that compress the cache during the pre-filling phase, with each press having a compression ratio attribute. The repository includes various training-free presses, special presses, and supports KV cache quantization. Users can contribute new presses and evaluate the performance of different presses on long-context datasets.
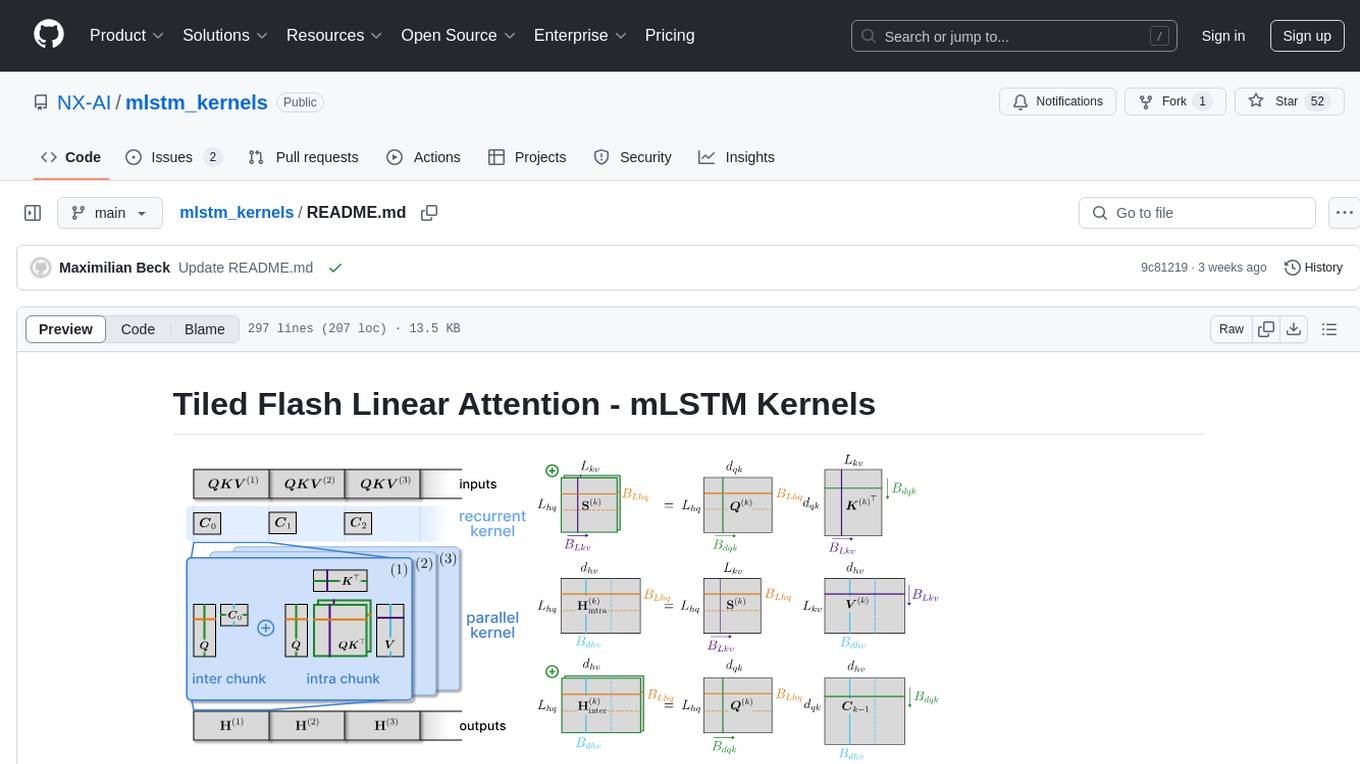
mlstm_kernels
This repository provides fast and efficient mLSTM training and inference Triton kernels built on Tiled Flash Linear Attention (TFLA). It includes implementations in JAX, PyTorch, and Triton, with chunkwise, parallel, and recurrent kernels for mLSTM. The repository also contains a benchmark library for runtime benchmarks and full mLSTM Huggingface models.
lantern
Lantern is an open-source PostgreSQL database extension designed to store vector data, generate embeddings, and handle vector search operations efficiently. It introduces a new index type called 'lantern_hnsw' for vector columns, which speeds up 'ORDER BY ... LIMIT' queries. Lantern utilizes the state-of-the-art HNSW implementation called usearch. Users can easily install Lantern using Docker, Homebrew, or precompiled binaries. The tool supports various distance functions, index construction parameters, and operator classes for efficient querying. Lantern offers features like embedding generation, interoperability with pgvector, parallel index creation, and external index graph generation. It aims to provide superior performance metrics compared to other similar tools and has a roadmap for future enhancements such as cloud-hosted version, hardware-accelerated distance metrics, industry-specific application templates, and support for version control and A/B testing of embeddings.
mimir
MIMIR is a Python package designed for measuring memorization in Large Language Models (LLMs). It provides functionalities for conducting experiments related to membership inference attacks on LLMs. The package includes implementations of various attacks such as Likelihood, Reference-based, Zlib Entropy, Neighborhood, Min-K% Prob, Min-K%++, Gradient Norm, and allows users to extend it by adding their own datasets and attacks.
llmgraph
llmgraph is a tool that enables users to create knowledge graphs in GraphML, GEXF, and HTML formats by extracting world knowledge from large language models (LLMs) like ChatGPT. It supports various entity types and relationships, offers cache support for efficient graph growth, and provides insights into LLM costs. Users can customize the model used and interact with different LLM providers. The tool allows users to generate interactive graphs based on a specified entity type and Wikipedia link, making it a valuable resource for knowledge graph creation and exploration.
Pixel-Reasoner
Pixel Reasoner is a framework that introduces reasoning in the pixel-space for Vision-Language Models (VLMs), enabling them to directly inspect, interrogate, and infer from visual evidences. This enhances reasoning fidelity for visual tasks by equipping VLMs with visual reasoning operations like zoom-in and select-frame. The framework addresses challenges like model's imbalanced competence and reluctance to adopt pixel-space operations through a two-phase training approach involving instruction tuning and curiosity-driven reinforcement learning. With these visual operations, VLMs can interact with complex visual inputs such as images or videos to gather necessary information, leading to improved performance across visual reasoning benchmarks.
sage
Sage is a tool that allows users to chat with any codebase, providing a chat interface for code understanding and integration. It simplifies the process of learning how a codebase works by offering heavily documented answers sourced directly from the code. Users can set up Sage locally or on the cloud with minimal effort. The tool is designed to be easily customizable, allowing users to swap components of the pipeline and improve the algorithms powering code understanding and generation.
LayerSkip
LayerSkip is an implementation enabling early exit inference and self-speculative decoding. It provides a code base for running models trained using the LayerSkip recipe, offering speedup through self-speculative decoding. The tool integrates with Hugging Face transformers and provides checkpoints for various LLMs. Users can generate tokens, benchmark on datasets, evaluate tasks, and sweep over hyperparameters to optimize inference speed. The tool also includes correctness verification scripts and Docker setup instructions. Additionally, other implementations like gpt-fast and Native HuggingFace are available. Training implementation is a work-in-progress, and contributions are welcome under the CC BY-NC license.
For similar tasks
langtrace
Langtrace is an open source observability software that lets you capture, debug, and analyze traces and metrics from all your applications that leverage LLM APIs, Vector Databases, and LLM-based Frameworks. It supports Open Telemetry Standards (OTEL), and the traces generated adhere to these standards. Langtrace offers both a managed SaaS version (Langtrace Cloud) and a self-hosted option. The SDKs for both Typescript/Javascript and Python are available, making it easy to integrate Langtrace into your applications. Langtrace automatically captures traces from various vendors, including OpenAI, Anthropic, Azure OpenAI, Langchain, LlamaIndex, Pinecone, and ChromaDB.
mlcraft
Synmetrix (prev. MLCraft) is an open source data engineering platform and semantic layer for centralized metrics management. It provides a complete framework for modeling, integrating, transforming, aggregating, and distributing metrics data at scale. Key features include data modeling and transformations, semantic layer for unified data model, scheduled reports and alerts, versioning, role-based access control, data exploration, caching, and collaboration on metrics modeling. Synmetrix leverages Cube (Cube.js) for flexible data models that consolidate metrics from various sources, enabling downstream distribution via a SQL API for integration into BI tools, reporting, dashboards, and data science. Use cases include data democratization, business intelligence, embedded analytics, and enhancing accuracy in data handling and queries. The tool speeds up data-driven workflows from metrics definition to consumption by combining data engineering best practices with self-service analytics capabilities.
synmetrix
Synmetrix is an open source data engineering platform and semantic layer for centralized metrics management. It provides a complete framework for modeling, integrating, transforming, aggregating, and distributing metrics data at scale. Key features include data modeling and transformations, semantic layer for unified data model, scheduled reports and alerts, versioning, role-based access control, data exploration, caching, and collaboration on metrics modeling. Synmetrix leverages Cube.js to consolidate metrics from various sources and distribute them downstream via a SQL API. Use cases include data democratization, business intelligence and reporting, embedded analytics, and enhancing accuracy in data handling and queries. The tool speeds up data-driven workflows from metrics definition to consumption by combining data engineering best practices with self-service analytics capabilities.

rtdl-num-embeddings
This repository provides the official implementation of the paper 'On Embeddings for Numerical Features in Tabular Deep Learning'. It focuses on transforming scalar continuous features into vectors before integrating them into the main backbone of tabular neural networks, showcasing improved performance. The embeddings for continuous features are shown to enhance the performance of tabular DL models and are applicable to various conventional backbones, offering efficiency comparable to Transformer-based models. The repository includes Python packages for practical usage, exploration of metrics and hyperparameters, and reproducing reported results for different algorithms and datasets.
VulBench
This repository contains materials for the paper 'How Far Have We Gone in Vulnerability Detection Using Large Language Model'. It provides a tool for evaluating vulnerability detection models using datasets such as d2a, ctf, magma, big-vul, and devign. Users can query the model 'Llama-2-7b-chat-hf' and store results in a SQLite database for analysis. The tool supports binary and multiple classification tasks with concurrency settings. Additionally, users can evaluate the results and generate a CSV file with metrics for each dataset and prompt type.
agentneo
AgentNeo is a Python package that provides functionalities for project, trace, dataset, experiment management. It allows users to authenticate, create projects, trace agents and LangGraph graphs, manage datasets, and run experiments with metrics. The tool aims to streamline AI project management and analysis by offering a comprehensive set of features.
hyperfy
Hyperfy is a powerful tool for automating social media marketing tasks. It provides a user-friendly interface to schedule posts, analyze performance metrics, and engage with followers across multiple platforms. With Hyperfy, users can save time and effort by streamlining their social media management processes in one centralized platform.
rill
Rill delivers rapid, self-service dashboards for data engineers and analysts directly on raw data lakes, ensuring reliable, fast-loading dashboards with accurate, real-time metrics. It comes with an embedded in-memory database for lightning-fast performance and supports bringing your own OLAP engine. Rill implements BI-as-code through SQL-based definitions, YAML configuration, Git integration, and CLI tools. Its metrics layer provides a unified way to define, compute, and serve business metrics, while AI agents can access fresh metrics instantly for precise decision-making and intelligent automation.
For similar jobs
sweep
Sweep is an AI junior developer that turns bugs and feature requests into code changes. It automatically handles developer experience improvements like adding type hints and improving test coverage.
teams-ai
The Teams AI Library is a software development kit (SDK) that helps developers create bots that can interact with Teams and Microsoft 365 applications. It is built on top of the Bot Framework SDK and simplifies the process of developing bots that interact with Teams' artificial intelligence capabilities. The SDK is available for JavaScript/TypeScript, .NET, and Python.
ai-guide
This guide is dedicated to Large Language Models (LLMs) that you can run on your home computer. It assumes your PC is a lower-end, non-gaming setup.

classifai
Supercharge WordPress Content Workflows and Engagement with Artificial Intelligence. Tap into leading cloud-based services like OpenAI, Microsoft Azure AI, Google Gemini and IBM Watson to augment your WordPress-powered websites. Publish content faster while improving SEO performance and increasing audience engagement. ClassifAI integrates Artificial Intelligence and Machine Learning technologies to lighten your workload and eliminate tedious tasks, giving you more time to create original content that matters.
chatbot-ui
Chatbot UI is an open-source AI chat app that allows users to create and deploy their own AI chatbots. It is easy to use and can be customized to fit any need. Chatbot UI is perfect for businesses, developers, and anyone who wants to create a chatbot.
BricksLLM
BricksLLM is a cloud native AI gateway written in Go. Currently, it provides native support for OpenAI, Anthropic, Azure OpenAI and vLLM. BricksLLM aims to provide enterprise level infrastructure that can power any LLM production use cases. Here are some use cases for BricksLLM: * Set LLM usage limits for users on different pricing tiers * Track LLM usage on a per user and per organization basis * Block or redact requests containing PIIs * Improve LLM reliability with failovers, retries and caching * Distribute API keys with rate limits and cost limits for internal development/production use cases * Distribute API keys with rate limits and cost limits for students
uAgents
uAgents is a Python library developed by Fetch.ai that allows for the creation of autonomous AI agents. These agents can perform various tasks on a schedule or take action on various events. uAgents are easy to create and manage, and they are connected to a fast-growing network of other uAgents. They are also secure, with cryptographically secured messages and wallets.
griptape
Griptape is a modular Python framework for building AI-powered applications that securely connect to your enterprise data and APIs. It offers developers the ability to maintain control and flexibility at every step. Griptape's core components include Structures (Agents, Pipelines, and Workflows), Tasks, Tools, Memory (Conversation Memory, Task Memory, and Meta Memory), Drivers (Prompt and Embedding Drivers, Vector Store Drivers, Image Generation Drivers, Image Query Drivers, SQL Drivers, Web Scraper Drivers, and Conversation Memory Drivers), Engines (Query Engines, Extraction Engines, Summary Engines, Image Generation Engines, and Image Query Engines), and additional components (Rulesets, Loaders, Artifacts, Chunkers, and Tokenizers). Griptape enables developers to create AI-powered applications with ease and efficiency.
