
cellseg_models.pytorch
Encoder-Decoder Cell and Nuclei segmentation models
Stars: 69
cellseg-models.pytorch is a Python library built upon PyTorch for 2D cell/nuclei instance segmentation models. It provides multi-task encoder-decoder architectures and post-processing methods for segmenting cell/nuclei instances. The library offers high-level API to define segmentation models, open-source datasets for training, flexibility to modify model components, sliding window inference, multi-GPU inference, benchmarking utilities, regularization techniques, and example notebooks for training and finetuning models with different backbones.
README:
cellseg-models.pytorch is a library built upon PyTorch that contains multi-task encoder-decoder architectures along with dedicated post-processing methods for segmenting cell/nuclei instances. As the name might suggest, this library is heavily inspired by segmentation_models.pytorch library for semantic segmentation.
- Now you can use any pre-trained image encoder from the timm library as the model backbone. (Given that they implement the
forward_intermediatesmethod, most of them do). - New example notebooks showing how to finetune Cellpose and Stardist with the new state-of-the-art foundation model backbones: UNI from the MahmoodLab, and Prov-GigaPath from Microsoft Research. Check out the notebooks here (UNI), and here (Prov-GigaPath).
- NOTE!: These foundation models are licensed under restrictive licences and you need to agree to the terms of any of the said models to get access to the weights. Once you have been granted access, you can run the above notebooks. You can request for access here: the model pages UNI and model pages Prov-GigaPath. These models may only be used for non-commercial, academic research purposes with proper attribution. Be sure that you have read and understood the terms before requesting access.
- High level API to define cell/nuclei instance segmentation models.
- 6 cell/nuclei instance segmentation model architectures and more to come.
- Open source datasets for training and benchmarking.
- Flexibility to modify the components of the model architectures.
- Sliding window inference for large images.
- Multi-GPU inference.
- All model architectures can be augmented to panoptic segmentation.
- Popular training losses and benchmarking metrics.
- Benchmarking utilities both for model latency & segmentation performance.
- Regularization techniques to tackle batch effects/domain shifts such as Strong Augment, Spectral decoupling, Label smoothing.
- Example notebooks to train models with lightning or accelerate.
- Example notebooks to finetune models with foundation model backbones such as UNI, Prov-GigaPath, and DINOv2.
pip install cellseg-models-pytorch| Model | Paper |
|---|---|
| [1] HoVer-Net | https://www.sciencedirect.com/science/article/pii/S1361841519301045?via%3Dihub |
| [2] Cellpose | https://www.nature.com/articles/s41592-020-01018-x |
| [3] Omnipose | https://www.biorxiv.org/content/10.1101/2021.11.03.467199v2 |
| [4] Stardist | https://arxiv.org/abs/1806.03535 |
| [5] CellVit-SAM | https://arxiv.org/abs/2306.15350.03535 |
| [6] CPP-Net | https://arxiv.org/abs/2102.0686703535 |
| Dataset | Paper |
|---|---|
| [7, 8] Pannuke | https://arxiv.org/abs/2003.10778 , https://link.springer.com/chapter/10.1007/978-3-030-23937-4_2 |
Finetuning CellPose with UNI backbone
-
Finetuning CellPose with UNI. Here we finetune the CellPose multi-class nuclei segmentation model with the foundation model
UNI-image-encoder backbone (checkout UNI). The Pannuke dataset (fold 1 & fold 2) are used for training data and the fold 3 is used as validation data. The model is trained (with checkpointing) by utilizing accelerate by hugginface. NOTE that you need to have granted access to the UNI weights and agreed to the terms of the model to be able to run the notebook.
Finetuning Stardist with Prov-GigaPath backbone
-
Finetuning Stardist with Prov-GigaPath. Here we finetune the Stardist multi-class nuclei segmentation model with the foundation model
Prov-GigaPath-image-encoder backbone (checkout Prov-GigaPath). The Pannuke dataset (fold 1 & fold 2) are used for training data and the fold 3 is used as validation data. The model is trained (with checkpointing) by utilizing accelerate by hugginface. NOTE that you need to have granted access to the Prov-GigaPath weights and agreed to the terms of the model to be able to run the notebook.
Finetuning CellPose with DINOv2 backbone
-
Finetuning CellPose with DINOv2 backbone for Pannuke. Here we finetune the CellPose multi-class nuclei segmentation model with a
LVD-142MpretrainedDINOv2backbone. The Pannuke dataset (fold 1 & fold 2) are used for training data and the fold 3 is used as validation data. The model is trained (with checkpointing) by utilizing lightning.
Finetuning CellVit-SAM with Pannuke
-
Finetuning CellVit-SAM with Pannuke. Here we finetune the CellVit-SAM multi-class nuclei segmentation model with a
SA-1BpretrainedSAM-image-encoder backbone (checkout SAM). The encoder is transformer basedVitDet-model. The Pannuke dataset (fold 1 & fold 2) are used for training data and the fold 3 is used as validation data. The model is trained (with checkpointing) by utilizing accelerate by hugginface.
Training Hover-Net with Pannuke
-
Training Hover-Net with Pannuke. Here we train the
Hover-Netnuclei segmentation model with animagenetpretrainedresnet50backbone from thetimmlibrary. The Pannuke dataset (fold 1 & fold 2) are used for training data and the fold 3 is used as validation data. The model is trained by utilizing lightning.
Training Stardist with Pannuke
-
Training Stardist with Pannuke. Here we train the
Stardistmulti-class nuclei segmentation model with animagenetpretrainedefficientnetv2_sbackbone from thetimmlibrary. The Pannuke dataset (fold 1 & fold 2) are used for training data and the fold 3 is used as validation data. The model is trained by utilizing lightning.
Training CellPose with Pannuke
-
Training CellPose with Pannuke. Here we train the
CellPosemulti-class nuclei segmentation model with animagenetpretrainedconvnext_smallbackbone from thetimmlibrary. The Pannuke dataset (fold 1 & fold 2) are used for training data and the fold 3 is used as validation data. The model is trained (with checkpointing) by utilizing accelerate by hugginface.
Training OmniPose with Pannuke
-
Training OmniPose with Pannuke. Here we train the OmniPose multi-class nuclei segmentation model with an
imagenetpretrainedfocalnet_small_lrfbackbone from thetimmlibrary. The Pannuke dataset (fold 1 & fold 2) are used for training data and the fold 3 is used as validation data. The model is trained (with checkpointing) by utilizing accelerate by hugginface.
Training CPP-Net with Pannuke
-
Training CPP-Net with Pannuke. Here we train the CPP-Net multi-class nuclei segmentation model with an
imagenetpretrainedefficientnetv2_sbackbone from thetimmlibrary. The Pannuke dataset (fold 1 & fold 2) are used for training data and the fold 3 is used as validation data. The model is trained by utilizing lightning.
Benchmarking Cellpose Trained on Pannuke
-
Benchmarking Cellpose Trained on Pannuke. Here we run benchmarking for
Cellposethat was trained on Pannuke. Both the model performance and latency benchmarking are covered.
Define Cellpose for cell segmentation.
import cellseg_models_pytorch as csmp
import torch
model = csmp.models.cellpose_base(type_classes=5)
x = torch.rand([1, 3, 256, 256])
# NOTE: the outputs still need post-processing.
y = model(x) # {"cellpose": [1, 2, 256, 256], "type": [1, 5, 256, 256]}Define Cellpose for cell and tissue area segmentation (Panoptic segmentation).
import cellseg_models_pytorch as csmp
import torch
model = csmp.models.cellpose_plus(type_classes=5, sem_classes=3)
x = torch.rand([1, 3, 256, 256])
# NOTE: the outputs still need post-processing.
y = model(x) # {"cellpose": [1, 2, 256, 256], "type": [1, 5, 256, 256], "sem": [1, 3, 256, 256]}Define panoptic Cellpose model with more flexibility.
import cellseg_models_pytorch as csmp
# the model will include two decoder branches.
decoders = ("cellpose", "sem")
# and in total three segmentation heads emerging from the decoders.
heads = {
"cellpose": {"cellpose": 2, "type": 5},
"sem": {"sem": 3}
}
model = csmp.CellPoseUnet(
decoders=decoders, # cellpose and semantic decoders
heads=heads, # three output heads
depth=5, # encoder depth
out_channels=(256, 128, 64, 32, 16), # num out channels at each decoder stage
layer_depths=(4, 4, 4, 4, 4), # num of conv blocks at each decoder layer
style_channels=256, # num of style vector channels
enc_name="resnet50", # timm encoder
enc_pretrain=True, # imagenet pretrained encoder
long_skip="unetpp", # unet++ long skips ("unet", "unetpp", "unet3p")
merge_policy="sum", # concatenate long skips ("cat", "sum")
short_skip="residual", # residual short skips ("basic", "residual", "dense")
normalization="bcn", # batch-channel-normalization.
activation="gelu", # gelu activation.
convolution="wsconv", # weight standardized conv.
attention="se", # squeeze-and-excitation attention.
pre_activate=False, # normalize and activation after convolution.
)
x = torch.rand([1, 3, 256, 256])
# NOTE: the outputs still need post-processing.
y = model(x) # {"cellpose": [1, 2, 256, 256], "type": [1, 5, 256, 256], "sem": [1, 3, 256, 256]}Run HoVer-Net inference and post-processing with a sliding window approach.
import cellseg_models_pytorch as csmp
# define the model
model = csmp.models.hovernet_base(type_classes=5)
# define the final activations for each model output
out_activations = {"hovernet": "tanh", "type": "softmax", "inst": "softmax"}
# define whether to weight down the predictions at the image boundaries
# typically, models perform the poorest at the image boundaries and with
# overlapping patches this causes issues which can be overcome by down-
# weighting the prediction boundaries
out_boundary_weights = {"hovernet": True, "type": False, "inst": False}
# define the inferer
inferer = csmp.inference.SlidingWindowInferer(
model=model,
input_folder="/path/to/images/",
checkpoint_path="/path/to/model/weights/",
out_activations=out_activations,
out_boundary_weights=out_boundary_weights,
instance_postproc="hovernet", # THE POST-PROCESSING METHOD
normalization="percentile", # same normalization as in training
patch_size=(256, 256),
stride=128,
padding=80,
batch_size=8,
)
inferer.infer()
inferer.out_masks
# {"image1" :{"inst": [H, W], "type": [H, W]}, ..., "imageN" :{"inst": [H, W], "type": [H, W]}}Generally, the model building API enables the effortless creation of hard-parameter sharing multi-task encoder-decoder CNN architectures. The general architectural schema is illustrated in the below image.
The class API enables the most flexibility in defining different model architectures. It borrows a lot from segmentation_models.pytorch models API.
Model classes:
csmp.CellPoseUnetcsmp.StarDistUnetcsmp.HoverNetcsmp.CellVitSAM
All of the models contain:
-
model.encoder- pretrained timm backbone for feature extraction. -
model.{decoder_name}_decoder- Models can have multiple decoders with unique names. -
model.{head_name}_seg_head- Model decoders can have multiple segmentation heads with unique names. -
model.forward(x)- forward pass. -
model.forward_features(x)- forward pass of the encoder and decoders. Returns enc and dec features
Defining your own multi-task architecture
For example, to define a multi-task architecture that has resnet50 encoder, four decoders, and 5 output heads with CellPoseUnet architectural components, we could do this:
import cellseg_models_pytorch as csmp
import torch
model = csmp.CellPoseUnet(
decoders=("cellpose", "dist", "contour", "sem"),
heads={
"cellpose": {"type": 5, "cellpose": 2},
"dist": {"dist": 1},
"contour": {"contour": 1},
"sem": {"sem": 4}
},
)
x = torch.rand([1, 3, 256, 256])
model(x)
# {
# "cellpose": [1, 2, 256, 256],
# "type": [1, 5, 256, 256],
# "dist": [1, 1, 256, 256],
# "contour": [1, 1, 256, 256],
# "sem": [1, 4, 256, 256]
# }With the function API, you can build models with low effort by calling the below listed functions. Under the hood, the function API simply calls the above classes with pre-defined decoder and head names. The training and post-processing tools of this library are built around these names, thus, it is recommended to use the function API, although, it is a bit more rigid than the class API. Basically, the function API only lacks the ability to define the output-tasks of the model, but allows for all the rest as the class API.
| Model functions | Output names | Task |
|---|---|---|
csmp.models.cellpose_base |
"type", "cellpose", |
instance segmentation |
csmp.models.cellpose_plus |
"type", "cellpose", "sem", |
panoptic segmentation |
csmp.models.omnipose_base |
"type", "omnipose"
|
instance segmentation |
csmp.models.omnipose_plus |
"type", "omnipose", "sem", |
panoptic segmentation |
csmp.models.hovernet_base |
"type", "inst", "hovernet"
|
instance segmentation |
csmp.models.hovernet_plus |
"type", "inst", "hovernet", "sem"
|
panoptic segmentation |
csmp.models.hovernet_small |
"type","hovernet"
|
instance segmentation |
csmp.models.hovernet_small_plus |
"type", "hovernet", "sem"
|
panoptic segmentation |
csmp.models.stardist_base |
"stardist", "dist"
|
binary instance segmentation |
csmp.models.stardist_base_multiclass |
"stardist", "dist", "type"
|
instance segmentation |
csmp.models.stardist_plus |
"stardist", "dist", "type", "sem"
|
panoptic segmentation |
csmp.models.cppnet_base |
"stardist_refined", "dist"
|
binary instance segmentation |
csmp.models.cppnet_base_multiclass |
"stardist_refined", "dist", "type"
|
instance segmentation |
csmp.models.cppnet_plus |
"stardist_refined", "dist", "type", "sem"
|
panoptic segmentation |
csmp.models.cellvit_sam_base |
"type", "inst", "hovernet"
|
instance segmentation |
csmp.models.cellvit_sam_plus |
"type", "inst", "hovernet", "sem"
|
panoptic segmentation |
csmp.models.cellvit_sam_small |
"type","hovernet"
|
instance segmentation |
csmp.models.cellvit_sam_small_plus |
"type", "hovernet", "sem"
|
panoptic segmentation |
- [1] S. Graham, Q. D. Vu, S. E. A. Raza, A. Azam, Y-W. Tsang, J. T. Kwak and N. Rajpoot. "HoVer-Net: Simultaneous Segmentation and Classification of Nuclei in Multi-Tissue Histology Images." Medical Image Analysis, Sept. 2019.
- [2] Stringer, C.; Wang, T.; Michaelos, M. & Pachitariu, M. Cellpose: a generalist algorithm for cellular segmentation Nature Methods, 2021, 18, 100-106
- [3] Cutler, K. J., Stringer, C., Wiggins, P. A., & Mougous, J. D. (2022). Omnipose: a high-precision morphology-independent solution for bacterial cell segmentation. bioRxiv. doi:10.1101/2021.11.03.467199
- [4] Uwe Schmidt, Martin Weigert, Coleman Broaddus, & Gene Myers (2018). Cell Detection with Star-Convex Polygons. In Medical Image Computing and Computer Assisted Intervention - MICCAI 2018 - 21st International Conference, Granada, Spain, September 16-20, 2018, Proceedings, Part II (pp. 265–273).
- [5] Hörst, F., Rempe, M., Heine, L., Seibold, C., Keyl, J., Baldini, G., Ugurel, S., Siveke, J., Grünwald, B., Egger, J., & Kleesiek, J. (2023). CellViT: Vision Transformers for Precise Cell Segmentation and Classification (Version 1). arXiv. https://doi.org/10.48550/ARXIV.2306.15350.
- [6] Chen, S., Ding, C., Liu, M., Cheng, J., & Tao, D. (2023). CPP-Net: Context-Aware Polygon Proposal Network for Nucleus Segmentation. In IEEE Transactions on Image Processing (Vol. 32, pp. 980–994). Institute of Electrical and Electronics Engineers (IEEE). https://doi.org/10.1109/tip.2023.3237013
- [7] Gamper, J., Koohbanani, N., Benet, K., Khuram, A., & Rajpoot, N. (2019) PanNuke: an open pan-cancer histology dataset for nuclei instance segmentation and classification. In European Congress on Digital Pathology (pp. 11-19).
- [8] Gamper, J., Koohbanani, N., Graham, S., Jahanifar, M., Khurram, S., Azam, A.,Hewitt, K., & Rajpoot, N. (2020). PanNuke Dataset Extension, Insights and Baselines. arXiv preprint arXiv:2003.10778.
- [9] Graham, S., Jahanifar, M., Azam, A., Nimir, M., Tsang, Y.W., Dodd, K., Hero, E., Sahota, H., Tank, A., Benes, K., & others (2021). Lizard: A Large-Scale Dataset for Colonic Nuclear Instance Segmentation and Classification. In Proceedings of the IEEE/CVF International Conference on Computer Vision (pp. 684-693).
@misc{csmp2022,
title={{cellseg_models.pytorch}: Cell/Nuclei Segmentation Models and Benchmark.},
author={Oskari Lehtonen},
howpublished = {\url{https://github.com/okunator/cellseg_models.pytorch}},
doi = {10.5281/zenodo.7064617}
year={2022}
}This project is distributed under MIT License
The project contains code from the original cell segmentation and 3rd-party libraries that have permissive licenses:
If you find this library useful in your project, it is your responsibility to ensure you comply with the conditions of any dependent licenses. Please create an issue if you think something is missing regarding the licenses.
For Tasks:
Click tags to check more tools for each tasksFor Jobs:
Alternative AI tools for cellseg_models.pytorch
Similar Open Source Tools

cellseg_models.pytorch
cellseg-models.pytorch is a Python library built upon PyTorch for 2D cell/nuclei instance segmentation models. It provides multi-task encoder-decoder architectures and post-processing methods for segmenting cell/nuclei instances. The library offers high-level API to define segmentation models, open-source datasets for training, flexibility to modify model components, sliding window inference, multi-GPU inference, benchmarking utilities, regularization techniques, and example notebooks for training and finetuning models with different backbones.
candle-vllm
Candle-vllm is an efficient and easy-to-use platform designed for inference and serving local LLMs, featuring an OpenAI compatible API server. It offers a highly extensible trait-based system for rapid implementation of new module pipelines, streaming support in generation, efficient management of key-value cache with PagedAttention, and continuous batching. The tool supports chat serving for various models and provides a seamless experience for users to interact with LLMs through different interfaces.
AnglE
AnglE is a library for training state-of-the-art BERT/LLM-based sentence embeddings with just a few lines of code. It also serves as a general sentence embedding inference framework, allowing for inferring a variety of transformer-based sentence embeddings. The library supports various loss functions such as AnglE loss, Contrastive loss, CoSENT loss, and Espresso loss. It provides backbones like BERT-based models, LLM-based models, and Bi-directional LLM-based models for training on single or multi-GPU setups. AnglE has achieved significant performance on various benchmarks and offers official pretrained models for both BERT-based and LLM-based models.
pixeltable
Pixeltable is a Python library designed for ML Engineers and Data Scientists to focus on exploration, modeling, and app development without the need to handle data plumbing. It provides a declarative interface for working with text, images, embeddings, and video, enabling users to store, transform, index, and iterate on data within a single table interface. Pixeltable is persistent, acting as a database unlike in-memory Python libraries such as Pandas. It offers features like data storage and versioning, combined data and model lineage, indexing, orchestration of multimodal workloads, incremental updates, and automatic production-ready code generation. The tool emphasizes transparency, reproducibility, cost-saving through incremental data changes, and seamless integration with existing Python code and libraries.
auto-round
AutoRound is an advanced weight-only quantization algorithm for low-bits LLM inference. It competes impressively against recent methods without introducing any additional inference overhead. The method adopts sign gradient descent to fine-tune rounding values and minmax values of weights in just 200 steps, often significantly outperforming SignRound with the cost of more tuning time for quantization. AutoRound is tailored for a wide range of models and consistently delivers noticeable improvements.
LLMTSCS
LLMLight is a novel framework that employs Large Language Models (LLMs) as decision-making agents for Traffic Signal Control (TSC). The framework leverages the advanced generalization capabilities of LLMs to engage in a reasoning and decision-making process akin to human intuition for effective traffic control. LLMLight has been demonstrated to be remarkably effective, generalizable, and interpretable against various transportation-based and RL-based baselines on nine real-world and synthetic datasets.
evalica
Evalica is a powerful tool for evaluating code quality and performance in software projects. It provides detailed insights and metrics to help developers identify areas for improvement and optimize their code. With support for multiple programming languages and frameworks, Evalica offers a comprehensive solution for code analysis and optimization. Whether you are a beginner looking to learn best practices or an experienced developer aiming to enhance your code quality, Evalica is the perfect tool for you.
rust-genai
genai is a multi-AI providers library for Rust that aims to provide a common and ergonomic single API to various generative AI providers such as OpenAI, Anthropic, Cohere, Ollama, and Gemini. It focuses on standardizing chat completion APIs across major AI services, prioritizing ergonomics and commonality. The library initially focuses on text chat APIs and plans to expand to support images, function calling, and more in the future versions. Version 0.1.x will have breaking changes in patches, while version 0.2.x will follow semver more strictly. genai does not provide a full representation of a given AI provider but aims to simplify the differences at a lower layer for ease of use.
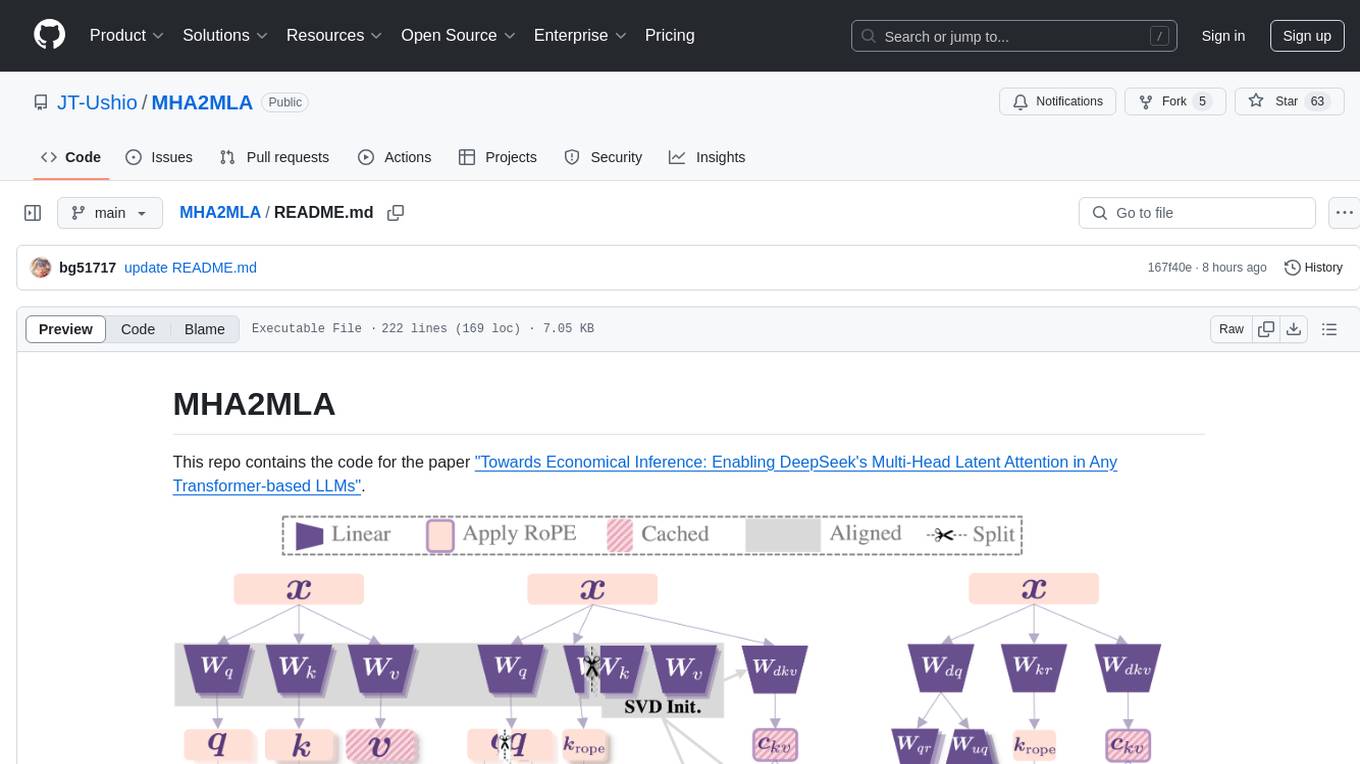
MHA2MLA
This repository contains the code for the paper 'Towards Economical Inference: Enabling DeepSeek's Multi-Head Latent Attention in Any Transformer-based LLMs'. It provides tools for fine-tuning and evaluating Llama models, converting models between different frameworks, processing datasets, and performing specific model training tasks like Partial-RoPE Fine-Tuning and Multiple-Head Latent Attention Fine-Tuning. The repository also includes commands for model evaluation using Lighteval and LongBench, along with necessary environment setup instructions.
TempCompass
TempCompass is a benchmark designed to evaluate the temporal perception ability of Video LLMs. It encompasses a diverse set of temporal aspects and task formats to comprehensively assess the capability of Video LLMs in understanding videos. The benchmark includes conflicting videos to prevent models from relying on single-frame bias and language priors. Users can clone the repository, install required packages, prepare data, run inference using examples like Video-LLaVA and Gemini, and evaluate the performance of their models across different tasks such as Multi-Choice QA, Yes/No QA, Caption Matching, and Caption Generation.
UHGEval
UHGEval is a comprehensive framework designed for evaluating the hallucination phenomena. It includes UHGEval, a framework for evaluating hallucination, XinhuaHallucinations dataset, and UHGEval-dataset pipeline for creating XinhuaHallucinations. The framework offers flexibility and extensibility for evaluating common hallucination tasks, supporting various models and datasets. Researchers can use the open-source pipeline to create customized datasets. Supported tasks include QA, dialogue, summarization, and multi-choice tasks.
ScaleLLM
ScaleLLM is a cutting-edge inference system engineered for large language models (LLMs), meticulously designed to meet the demands of production environments. It extends its support to a wide range of popular open-source models, including Llama3, Gemma, Bloom, GPT-NeoX, and more. ScaleLLM is currently undergoing active development. We are fully committed to consistently enhancing its efficiency while also incorporating additional features. Feel free to explore our **_Roadmap_** for more details. ## Key Features * High Efficiency: Excels in high-performance LLM inference, leveraging state-of-the-art techniques and technologies like Flash Attention, Paged Attention, Continuous batching, and more. * Tensor Parallelism: Utilizes tensor parallelism for efficient model execution. * OpenAI-compatible API: An efficient golang rest api server that compatible with OpenAI. * Huggingface models: Seamless integration with most popular HF models, supporting safetensors. * Customizable: Offers flexibility for customization to meet your specific needs, and provides an easy way to add new models. * Production Ready: Engineered with production environments in mind, ScaleLLM is equipped with robust system monitoring and management features to ensure a seamless deployment experience.
VLM-R1
VLM-R1 is a stable and generalizable R1-style Large Vision-Language Model proposed for Referring Expression Comprehension (REC) task. It compares R1 and SFT approaches, showing R1 model's steady improvement on out-of-domain test data. The project includes setup instructions, training steps for GRPO and SFT models, support for user data loading, and evaluation process. Acknowledgements to various open-source projects and resources are mentioned. The project aims to provide a reliable and versatile solution for vision-language tasks.

CodeTF
CodeTF is a Python transformer-based library for code large language models (Code LLMs) and code intelligence. It provides an interface for training and inferencing on tasks like code summarization, translation, and generation. The library offers utilities for code manipulation across various languages, including easy extraction of code attributes. Using tree-sitter as its core AST parser, CodeTF enables parsing of function names, comments, and variable names. It supports fast model serving, fine-tuning of LLMs, various code intelligence tasks, preprocessed datasets, model evaluation, pretrained and fine-tuned models, and utilities to manipulate source code. CodeTF aims to facilitate the integration of state-of-the-art Code LLMs into real-world applications, ensuring a user-friendly environment for code intelligence tasks.
openlrc
Open-Lyrics is a Python library that transcribes voice files using faster-whisper and translates/polishes the resulting text into `.lrc` files in the desired language using LLM, e.g. OpenAI-GPT, Anthropic-Claude. It offers well preprocessed audio to reduce hallucination and context-aware translation to improve translation quality. Users can install the library from PyPI or GitHub and follow the installation steps to set up the environment. The tool supports GUI usage and provides Python code examples for transcription and translation tasks. It also includes features like utilizing context and glossary for translation enhancement, pricing information for different models, and a list of todo tasks for future improvements.
Apollo
Apollo is a multilingual medical LLM that covers English, Chinese, French, Hindi, Spanish, Hindi, and Arabic. It is designed to democratize medical AI to 6B people. Apollo has achieved state-of-the-art results on a variety of medical NLP tasks, including question answering, medical dialogue generation, and medical text classification. Apollo is easy to use and can be integrated into a variety of applications, making it a valuable tool for healthcare professionals and researchers.
For similar tasks

cellseg_models.pytorch
cellseg-models.pytorch is a Python library built upon PyTorch for 2D cell/nuclei instance segmentation models. It provides multi-task encoder-decoder architectures and post-processing methods for segmenting cell/nuclei instances. The library offers high-level API to define segmentation models, open-source datasets for training, flexibility to modify model components, sliding window inference, multi-GPU inference, benchmarking utilities, regularization techniques, and example notebooks for training and finetuning models with different backbones.
pyllms
PyLLMs is a minimal Python library designed to connect to various Language Model Models (LLMs) such as OpenAI, Anthropic, Google, AI21, Cohere, Aleph Alpha, and HuggingfaceHub. It provides a built-in model performance benchmark for fast prototyping and evaluating different models. Users can easily connect to top LLMs, get completions from multiple models simultaneously, and evaluate models on quality, speed, and cost. The library supports asynchronous completion, streaming from compatible models, and multi-model initialization for testing and comparison. Additionally, it offers features like passing chat history, system messages, counting tokens, and benchmarking models based on quality, speed, and cost.
LLM-Fine-Tuning-Azure
A fine-tuning guide for both OpenAI and Open-Source Large Language Models on Azure. Fine-Tuning retrains an existing pre-trained LLM using example data, resulting in a new 'custom' fine-tuned LLM optimized for task-specific examples. Use cases include improving LLM performance on specific tasks and introducing information not well represented by the base LLM model. Suitable for cases where latency is critical, high accuracy is required, and clear evaluation metrics are available. Learning path includes labs for fine-tuning GPT and Llama2 models via Dashboards and Python SDK.
awesome-mobile-llm
Awesome Mobile LLMs is a curated list of Large Language Models (LLMs) and related studies focused on mobile and embedded hardware. The repository includes information on various LLM models, deployment frameworks, benchmarking efforts, applications, multimodal LLMs, surveys on efficient LLMs, training LLMs on device, mobile-related use-cases, industry announcements, and related repositories. It aims to be a valuable resource for researchers, engineers, and practitioners interested in mobile LLMs.
Medical_Image_Analysis
The Medical_Image_Analysis repository focuses on X-ray image-based medical report generation using large language models. It provides pre-trained models and benchmarks for CheXpert Plus dataset, context sample retrieval for X-ray report generation, and pre-training on high-definition X-ray images. The goal is to enhance diagnostic accuracy and reduce patient wait times by improving X-ray report generation through advanced AI techniques.

AngelSlim
AngelSlim is a comprehensive and efficient large model compression toolkit designed to be user-friendly. It integrates mainstream compression algorithms for easy one-click access, continuously innovates compression algorithms, and optimizes end-to-end performance in model compression and deployment. It supports various models for quantization and speculative sampling, with a focus on performance optimization and ease of use.
For similar jobs

cellseg_models.pytorch
cellseg-models.pytorch is a Python library built upon PyTorch for 2D cell/nuclei instance segmentation models. It provides multi-task encoder-decoder architectures and post-processing methods for segmenting cell/nuclei instances. The library offers high-level API to define segmentation models, open-source datasets for training, flexibility to modify model components, sliding window inference, multi-GPU inference, benchmarking utilities, regularization techniques, and example notebooks for training and finetuning models with different backbones.
aicsimageio
AICSImageIO is a Python tool for Image Reading, Metadata Conversion, and Image Writing for Microscopy Images. It supports various file formats like OME-TIFF, TIFF, ND2, DV, CZI, LIF, PNG, GIF, and Bio-Formats. Users can read and write metadata and imaging data, work with different file systems like local paths, HTTP URLs, s3fs, and gcsfs. The tool provides functionalities for full image reading, delayed image reading, mosaic image reading, metadata reading, xarray coordinate plane attachment, cloud IO support, and saving to OME-TIFF. It also offers benchmarking and developer resources.

KG_RAG
KG-RAG (Knowledge Graph-based Retrieval Augmented Generation) is a task agnostic framework that combines the explicit knowledge of a Knowledge Graph (KG) with the implicit knowledge of a Large Language Model (LLM). KG-RAG extracts "prompt-aware context" from a KG, which is defined as the minimal context sufficient enough to respond to the user prompt. This framework empowers a general-purpose LLM by incorporating an optimized domain-specific 'prompt-aware context' from a biomedical KG. KG-RAG is specifically designed for running prompts related to Diseases.

Scientific-LLM-Survey
Scientific Large Language Models (Sci-LLMs) is a repository that collects papers on scientific large language models, focusing on biology and chemistry domains. It includes textual, molecular, protein, and genomic languages, as well as multimodal language. The repository covers various large language models for tasks such as molecule property prediction, interaction prediction, protein sequence representation, protein sequence generation/design, DNA-protein interaction prediction, and RNA prediction. It also provides datasets and benchmarks for evaluating these models. The repository aims to facilitate research and development in the field of scientific language modeling.
biochatter
Generative AI models have shown tremendous usefulness in increasing accessibility and automation of a wide range of tasks. This repository contains the `biochatter` Python package, a generic backend library for the connection of biomedical applications to conversational AI. It aims to provide a common framework for deploying, testing, and evaluating diverse models and auxiliary technologies in the biomedical domain. BioChatter is part of the BioCypher ecosystem, connecting natively to BioCypher knowledge graphs.
ceLLama
ceLLama is a streamlined automation pipeline for cell type annotations using large-language models (LLMs). It operates locally to ensure privacy, provides comprehensive analysis by considering negative genes, offers efficient processing speed, and generates customized reports. Ideal for quick and preliminary cell type checks.
PINNACLE
PINNACLE is a flexible geometric deep learning approach that trains on contextualized protein interaction networks to generate context-aware protein representations. It provides protein representations split across various cell-type contexts from different tissues and organs. The tool can be fine-tuned to study the genomic effects of drugs and nominate promising protein targets and cell-type contexts for further investigation. PINNACLE exemplifies the paradigm of incorporating context-specific effects for studying biological systems, especially the impact of disease and therapeutics.
Taiyi-LLM
Taiyi (太一) is a bilingual large language model fine-tuned for diverse biomedical tasks. It aims to facilitate communication between healthcare professionals and patients, provide medical information, and assist in diagnosis, biomedical knowledge discovery, drug development, and personalized healthcare solutions. The model is based on the Qwen-7B-base model and has been fine-tuned using rich bilingual instruction data. It covers tasks such as question answering, biomedical dialogue, medical report generation, biomedical information extraction, machine translation, title generation, text classification, and text semantic similarity. The project also provides standardized data formats, model training details, model inference guidelines, and overall performance metrics across various BioNLP tasks.







