aicsimageio
Image Reading, Metadata Conversion, and Image Writing for Microscopy Images in Python
Stars: 198
AICSImageIO is a Python tool for Image Reading, Metadata Conversion, and Image Writing for Microscopy Images. It supports various file formats like OME-TIFF, TIFF, ND2, DV, CZI, LIF, PNG, GIF, and Bio-Formats. Users can read and write metadata and imaging data, work with different file systems like local paths, HTTP URLs, s3fs, and gcsfs. The tool provides functionalities for full image reading, delayed image reading, mosaic image reading, metadata reading, xarray coordinate plane attachment, cloud IO support, and saving to OME-TIFF. It also offers benchmarking and developer resources.
README:
[!WARNING]
AICSImageIO is now in maintenance mode only. Please take a look at its compatible successorbioio(see here for migration guide)
Image Reading, Metadata Conversion, and Image Writing for Microscopy Images in Pure Python
-
Supports reading metadata and imaging data for:
OME-TIFFTIFF-
ND2-- (pip install aicsimageio[nd2]) -
DV-- (pip install aicsimageio[dv]) -
CZI-- (pip install aicspylibczi>=3.1.1 fsspec>=2022.8.0) -
LIF-- (pip install readlif>=0.6.4) -
PNG,GIF, etc. -- (pip install aicsimageio[base-imageio]) - Files supported by Bio-Formats -- (
pip install aicsimageio bioformats_jar) (Note: requiresjavaandmaven, see below for details.)
-
Supports writing metadata and imaging data for:
OME-TIFF-
PNG,GIF, etc. -- (pip install aicsimageio[base-imageio])
-
Supports reading and writing to fsspec supported file systems wherever possible:
- Local paths (i.e.
my-file.png) - HTTP URLs (i.e.
https://my-domain.com/my-file.png) -
s3fs (i.e.
s3://my-bucket/my-file.png) -
gcsfs (i.e.
gcs://my-bucket/my-file.png)
See Cloud IO Support for more details.
- Local paths (i.e.
Stable Release: pip install aicsimageio
Development Head: pip install git+https://github.com/AllenCellModeling/aicsimageio.git
AICSImageIO is supported on Windows, Mac, and Ubuntu. For other platforms, you will likely need to build from source.
TIFF and OME-TIFF reading and writing is always available after
installing aicsimageio, but extra supported formats can be
optionally installed using [...] syntax.
- For a single additional supported format (e.g. ND2):
pip install aicsimageio[nd2] - For a single additional supported format (e.g. ND2), development head:
pip install "aicsimageio[nd2] @ git+https://github.com/AllenCellModeling/aicsimageio.git" - For a single additional supported format (e.g. ND2), specific tag (e.g.
v4.0.0.dev6):pip install "aicsimageio[nd2] @ git+https://github.com/AllenCellModeling/[email protected]" - For faster OME-TIFF reading with tile tags:
pip install aicsimageio[bfio] - For multiple additional supported formats:
pip install aicsimageio[base-imageio,nd2] - For all additional supported (and openly licensed) formats:
pip install aicsimageio[all] - Due to the GPL license, LIF support is not included with the
[all]extra, and must be installed manually withpip install aicsimageio readlif>=0.6.4 - Due to the GPL license, CZI support is not included with the
[all]extra, and must be installed manually withpip install aicsimageio aicspylibczi>=3.1.1 fsspec>=2022.8.0 - Due to the GPL license, Bio-Formats support is not included with the
[all]extra, and must be installed manually withpip install aicsimageio bioformats_jar. Important!! Bio-Formats support also requires ajavaandmvnexecutable in the environment. The simplest method is to installbioformats_jarfrom conda:conda install -c conda-forge bioformats_jar(which will additionally bringopenjdkandmavenpackages).
For full package documentation please visit allencellmodeling.github.io/aicsimageio.
If your image fits in memory:
from aicsimageio import AICSImage
# Get an AICSImage object
img = AICSImage("my_file.tiff") # selects the first scene found
img.data # returns 5D TCZYX numpy array
img.xarray_data # returns 5D TCZYX xarray data array backed by numpy
img.dims # returns a Dimensions object
img.dims.order # returns string "TCZYX"
img.dims.X # returns size of X dimension
img.shape # returns tuple of dimension sizes in TCZYX order
img.get_image_data("CZYX", T=0) # returns 4D CZYX numpy array
# Get the id of the current operating scene
img.current_scene
# Get a list valid scene ids
img.scenes
# Change scene using name
img.set_scene("Image:1")
# Or by scene index
img.set_scene(1)
# Use the same operations on a different scene
# ...The .data and .xarray_data properties will load the whole scene into memory.
The .get_image_data function will load the whole scene into memory and then retrieve
the specified chunk.
If your image doesn't fit in memory:
from aicsimageio import AICSImage
# Get an AICSImage object
img = AICSImage("my_file.tiff") # selects the first scene found
img.dask_data # returns 5D TCZYX dask array
img.xarray_dask_data # returns 5D TCZYX xarray data array backed by dask array
img.dims # returns a Dimensions object
img.dims.order # returns string "TCZYX"
img.dims.X # returns size of X dimension
img.shape # returns tuple of dimension sizes in TCZYX order
# Pull only a specific chunk in-memory
lazy_t0 = img.get_image_dask_data("CZYX", T=0) # returns out-of-memory 4D dask array
t0 = lazy_t0.compute() # returns in-memory 4D numpy array
# Get the id of the current operating scene
img.current_scene
# Get a list valid scene ids
img.scenes
# Change scene using name
img.set_scene("Image:1")
# Or by scene index
img.set_scene(1)
# Use the same operations on a different scene
# ...The .dask_data and .xarray_dask_data properties and the .get_image_dask_data
function will not load any piece of the imaging data into memory until you specifically
call .compute on the returned Dask array. In doing so, you will only then load the
selected chunk in-memory.
Read stitched data or single tiles as a dimension.
Readers that support mosaic tile stitching:
LifReaderCziReader
If the file format reader supports stitching mosaic tiles together, the
AICSImage object will default to stitching the tiles back together.
img = AICSImage("very-large-mosaic.lif")
img.dims.order # T, C, Z, big Y, big X, (S optional)
img.dask_data # Dask chunks fall on tile boundaries, pull YX chunks out of the imageThis behavior can be manually turned off:
img = AICSImage("very-large-mosaic.lif", reconstruct_mosaic=False)
img.dims.order # M (tile index), T, C, Z, small Y, small X, (S optional)
img.dask_data # Chunks use normal ZYXIf the reader does not support stitching tiles together the M tile index will be
available on the AICSImage object:
img = AICSImage("some-unsupported-mosaic-stitching-format.ext")
img.dims.order # M (tile index), T, C, Z, small Y, small X, (S optional)
img.dask_data # Chunks use normal ZYXIf the file format reader detects mosaic tiles in the image, the Reader object
will store the tiles as a dimension.
If tile stitching is implemented, the Reader can also return the stitched image.
reader = LifReader("ver-large-mosaic.lif")
reader.dims.order # M, T, C, Z, tile size Y, tile size X, (S optional)
reader.dask_data # normal operations, can use M dimension to select individual tiles
reader.mosaic_dask_data # returns stitched mosaic - T, C, Z, big Y, big, X, (S optional)There are functions available on both the AICSImage and Reader objects
to help with single tile positioning:
img = AICSImage("very-large-mosaic.lif")
img.mosaic_tile_dims # Returns a Dimensions object with just Y and X dim sizes
img.mosaic_tile_dims.Y # 512 (for example)
# Get the tile start indices (top left corner of tile)
y_start_index, x_start_index = img.get_mosaic_tile_position(12)from aicsimageio import AICSImage
# Get an AICSImage object
img = AICSImage("my_file.tiff") # selects the first scene found
img.metadata # returns the metadata object for this file format (XML, JSON, etc.)
img.channel_names # returns a list of string channel names found in the metadata
img.physical_pixel_sizes.Z # returns the Z dimension pixel size as found in the metadata
img.physical_pixel_sizes.Y # returns the Y dimension pixel size as found in the metadata
img.physical_pixel_sizes.X # returns the X dimension pixel size as found in the metadataIf aicsimageio finds coordinate information for the spatial-temporal dimensions of
the image in metadata, you can use
xarray for indexing by coordinates.
from aicsimageio import AICSImage
# Get an AICSImage object
img = AICSImage("my_file.ome.tiff")
# Get the first ten seconds (not frames)
first_ten_seconds = img.xarray_data.loc[:10] # returns an xarray.DataArray
# Get the first ten major units (usually micrometers, not indices) in Z
first_ten_mm_in_z = img.xarray_data.loc[:, :, :10]
# Get the first ten major units (usually micrometers, not indices) in Y
first_ten_mm_in_y = img.xarray_data.loc[:, :, :, :10]
# Get the first ten major units (usually micrometers, not indices) in X
first_ten_mm_in_x = img.xarray_data.loc[:, :, :, :, :10]See xarray
"Indexing and Selecting Data" Documentation
for more information.
File-System Specification (fsspec) allows for common object storage services (S3, GCS, etc.) to act like normal filesystems by following the same base specification across them all. AICSImageIO utilizes this standard specification to make it possible to read directly from remote resources when the specification is installed.
from aicsimageio import AICSImage
# Get an AICSImage object
img = AICSImage("http://my-website.com/my_file.tiff")
img = AICSImage("s3://my-bucket/my_file.tiff")
img = AICSImage("gcs://my-bucket/my_file.tiff")
# Or read with specific filesystem creation arguments
img = AICSImage("s3://my-bucket/my_file.tiff", fs_kwargs=dict(anon=True))
img = AICSImage("gcs://my-bucket/my_file.tiff", fs_kwargs=dict(anon=True))
# All other normal operations work just fineRemote reading requires that the file-system specification implementation for the target backend is installed.
- For
s3:pip install s3fs - For
gs:pip install gcsfs
See the list of known implementations.
The simpliest method to save your image as an OME-TIFF file with key pieces of
metadata is to use the save function.
from aicsimageio import AICSImage
AICSImage("my_file.czi").save("my_file.ome.tiff")Note: By default aicsimageio will generate only a portion of metadata to pass
along from the reader to the OME model. This function currently does not do a full
metadata translation.
For finer grain customization of the metadata, scenes, or if you want to save an array as an OME-TIFF, the writer class can also be used to customize as needed.
import numpy as np
from aicsimageio.writers import OmeTiffWriter
image = np.random.rand(10, 3, 1024, 2048)
OmeTiffWriter.save(image, "file.ome.tif", dim_order="ZCYX")See OmeTiffWriter documentation for more details.
In most cases, AICSImage.save is usually a good default but there are other image
writers available. For more information, please refer to
our writers documentation.
AICSImageIO is benchmarked using asv.
You can find the benchmark results for every commit to main starting at the 4.0
release on our
benchmarks page.
See our developer resources for information related to developing the code.
If you find aicsimageio useful, please cite this repository as:
Eva Maxfield Brown, Dan Toloudis, Jamie Sherman, Madison Swain-Bowden, Talley Lambert, AICSImageIO Contributors (2021). AICSImageIO: Image Reading, Metadata Conversion, and Image Writing for Microscopy Images in Pure Python [Computer software]. GitHub. https://github.com/AllenCellModeling/aicsimageio
bibtex:
@misc{aicsimageio,
author = {Brown, Eva Maxfield and Toloudis, Dan and Sherman, Jamie and Swain-Bowden, Madison and Lambert, Talley and {AICSImageIO Contributors}},
title = {AICSImageIO: Image Reading, Metadata Conversion, and Image Writing for Microscopy Images in Pure Python},
year = {2021},
publisher = {GitHub},
url = {https://github.com/AllenCellModeling/aicsimageio}
}Free software: BSD-3-Clause
(The LIF component is licensed under GPLv3 and is not included in this package) (The Bio-Formats component is licensed under GPLv2 and is not included in this package) (The CZI component is licensed under GPLv3 and is not included in this package)
For Tasks:
Click tags to check more tools for each tasksFor Jobs:
Alternative AI tools for aicsimageio
Similar Open Source Tools

aicsimageio
AICSImageIO is a Python tool for Image Reading, Metadata Conversion, and Image Writing for Microscopy Images. It supports various file formats like OME-TIFF, TIFF, ND2, DV, CZI, LIF, PNG, GIF, and Bio-Formats. Users can read and write metadata and imaging data, work with different file systems like local paths, HTTP URLs, s3fs, and gcsfs. The tool provides functionalities for full image reading, delayed image reading, mosaic image reading, metadata reading, xarray coordinate plane attachment, cloud IO support, and saving to OME-TIFF. It also offers benchmarking and developer resources.

raglite
RAGLite is a Python toolkit for Retrieval-Augmented Generation (RAG) with PostgreSQL or SQLite. It offers configurable options for choosing LLM providers, database types, and rerankers. The toolkit is fast and permissive, utilizing lightweight dependencies and hardware acceleration. RAGLite provides features like PDF to Markdown conversion, multi-vector chunk embedding, optimal semantic chunking, hybrid search capabilities, adaptive retrieval, and improved output quality. It is extensible with a built-in Model Context Protocol server, customizable ChatGPT-like frontend, document conversion to Markdown, and evaluation tools. Users can configure RAGLite for various tasks like configuring, inserting documents, running RAG pipelines, computing query adapters, evaluating performance, running MCP servers, and serving frontends.
hqq
HQQ is a fast and accurate model quantizer that skips the need for calibration data. It's super simple to implement (just a few lines of code for the optimizer). It can crunch through quantizing the Llama2-70B model in only 4 minutes! 🚀
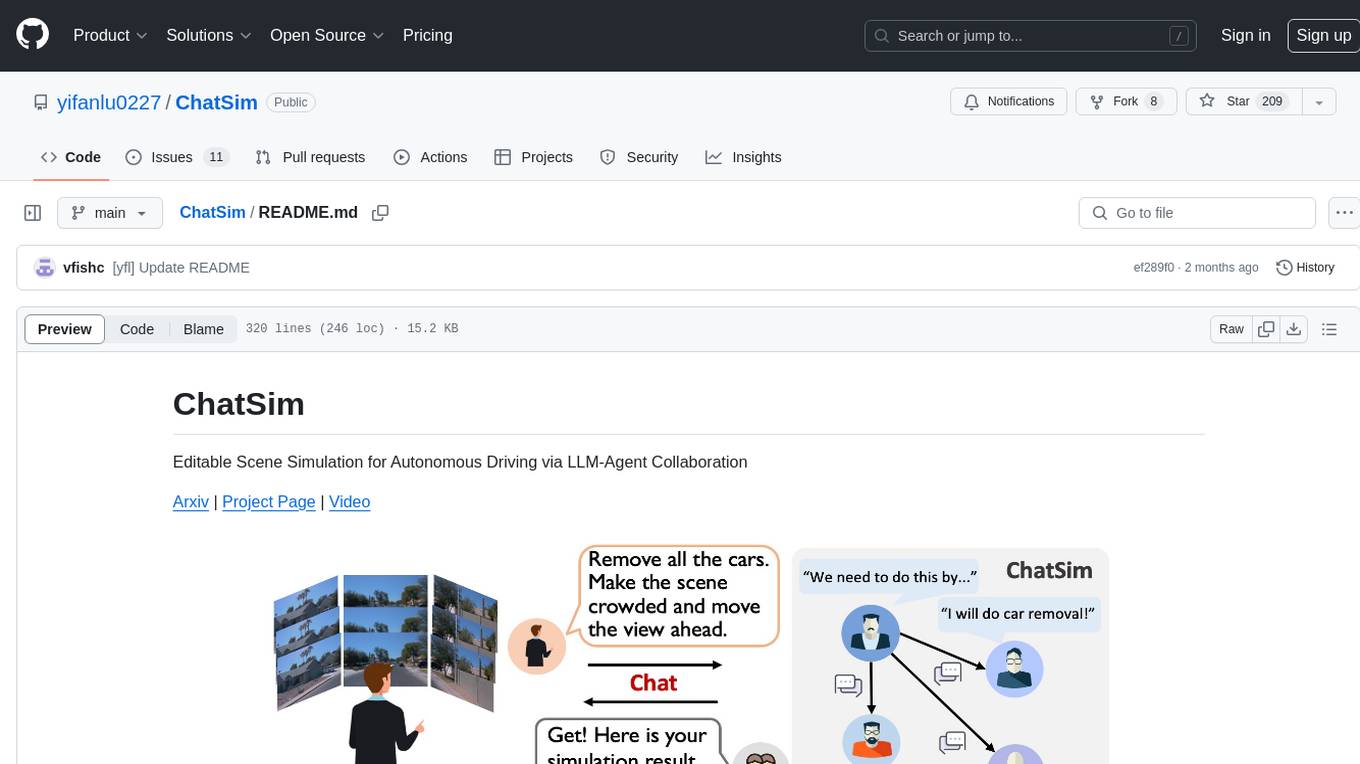
ChatSim
ChatSim is a tool designed for editable scene simulation for autonomous driving via LLM-Agent collaboration. It provides functionalities for setting up the environment, installing necessary dependencies like McNeRF and Inpainting tools, and preparing data for simulation. Users can train models, simulate scenes, and track trajectories for smoother and more realistic results. The tool integrates with Blender software and offers options for training McNeRF models and McLight's skydome estimation network. It also includes a trajectory tracking module for improved trajectory tracking. ChatSim aims to facilitate the simulation of autonomous driving scenarios with collaborative LLM-Agents.

shellChatGPT
ShellChatGPT is a shell wrapper for OpenAI's ChatGPT, DALL-E, Whisper, and TTS, featuring integration with LocalAI, Ollama, Gemini, Mistral, Groq, and GitHub Models. It provides text and chat completions, vision, reasoning, and audio models, voice-in and voice-out chatting mode, text editor interface, markdown rendering support, session management, instruction prompt manager, integration with various service providers, command line completion, file picker dialogs, color scheme personalization, stdin and text file input support, and compatibility with Linux, FreeBSD, MacOS, and Termux for a responsive experience.
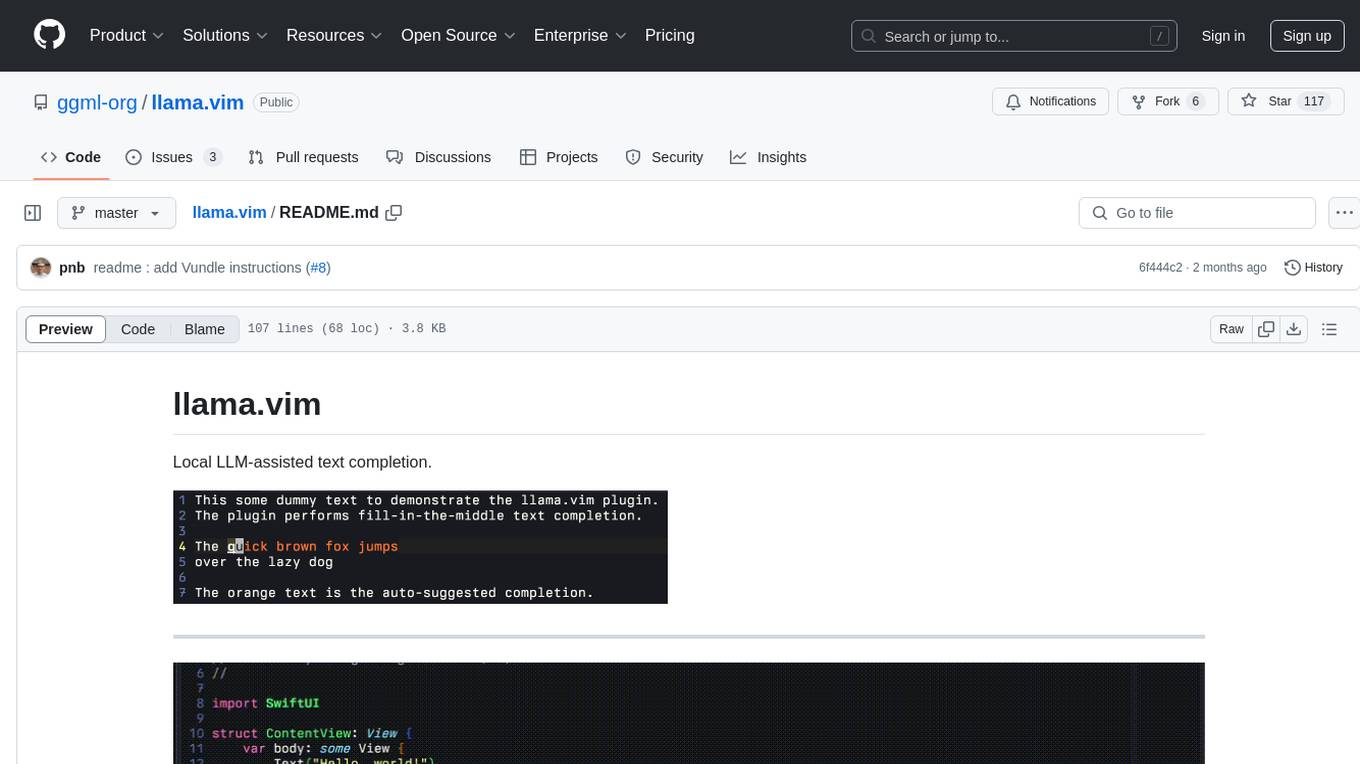
llama.vim
llama.vim is a plugin that provides local LLM-assisted text completion for Vim users. It offers features such as auto-suggest on cursor movement, manual suggestion toggling, suggestion acceptance with Tab and Shift+Tab, control over text generation time, context configuration, ring context with chunks from open and edited files, and performance stats display. The plugin requires a llama.cpp server instance to be running and supports FIM-compatible models. It aims to be simple, lightweight, and provide high-quality and performant local FIM completions even on consumer-grade hardware.
react-native-fast-tflite
A high-performance TensorFlow Lite library for React Native that utilizes JSI for power, zero-copy ArrayBuffers for efficiency, and low-level C/C++ TensorFlow Lite core API for direct memory access. It supports swapping out TensorFlow Models at runtime and GPU-accelerated delegates like CoreML/Metal/OpenGL. Easy VisionCamera integration allows for seamless usage. Users can load TensorFlow Lite models, interpret input and output data, and utilize GPU Delegates for faster computation. The library is suitable for real-time object detection, image classification, and other machine learning tasks in React Native applications.
models.dev
Models.dev is an open-source database providing detailed specifications, pricing, and capabilities of various AI models. It serves as a centralized platform for accessing information on AI models, allowing users to contribute and utilize the data through an API. The repository contains data stored in TOML files, organized by provider and model, along with SVG logos. Users can contribute by adding new models following specific guidelines and submitting pull requests for validation. The project aims to maintain an up-to-date and comprehensive database of AI model information.
stark
STaRK is a large-scale semi-structure retrieval benchmark on Textual and Relational Knowledge Bases. It provides natural-sounding and practical queries crafted to incorporate rich relational information and complex textual properties, closely mirroring real-life scenarios. The benchmark aims to assess how effectively large language models can handle the interplay between textual and relational requirements in queries, using three diverse knowledge bases constructed from public sources.
paxml
Pax is a framework to configure and run machine learning experiments on top of Jax.

wllama
Wllama is a WebAssembly binding for llama.cpp, a high-performance and lightweight language model library. It enables you to run inference directly on the browser without the need for a backend or GPU. Wllama provides both high-level and low-level APIs, allowing you to perform various tasks such as completions, embeddings, tokenization, and more. It also supports model splitting, enabling you to load large models in parallel for faster download. With its Typescript support and pre-built npm package, Wllama is easy to integrate into your React Typescript projects.
openai
An open-source client package that allows developers to easily integrate the power of OpenAI's state-of-the-art AI models into their Dart/Flutter applications. The library provides simple and intuitive methods for making requests to OpenAI's various APIs, including the GPT-3 language model, DALL-E image generation, and more. It is designed to be lightweight and easy to use, enabling developers to focus on building their applications without worrying about the complexities of dealing with HTTP requests. Note that this is an unofficial library as OpenAI does not have an official Dart library.
langserve
LangServe helps developers deploy `LangChain` runnables and chains as a REST API. This library is integrated with FastAPI and uses pydantic for data validation. In addition, it provides a client that can be used to call into runnables deployed on a server. A JavaScript client is available in LangChain.js.
neural-speed
Neural Speed is an innovative library designed to support the efficient inference of large language models (LLMs) on Intel platforms through the state-of-the-art (SOTA) low-bit quantization powered by Intel Neural Compressor. The work is inspired by llama.cpp and further optimized for Intel platforms with our innovations in NeurIPS' 2023
metis
Metis is an open-source, AI-driven tool for deep security code review, created by Arm's Product Security Team. It helps engineers detect subtle vulnerabilities, improve secure coding practices, and reduce review fatigue. Metis uses LLMs for semantic understanding and reasoning, RAG for context-aware reviews, and supports multiple languages and vector store backends. It provides a plugin-friendly and extensible architecture, named after the Greek goddess of wisdom, Metis. The tool is designed for large, complex, or legacy codebases where traditional tooling falls short.
For similar tasks

aicsimageio
AICSImageIO is a Python tool for Image Reading, Metadata Conversion, and Image Writing for Microscopy Images. It supports various file formats like OME-TIFF, TIFF, ND2, DV, CZI, LIF, PNG, GIF, and Bio-Formats. Users can read and write metadata and imaging data, work with different file systems like local paths, HTTP URLs, s3fs, and gcsfs. The tool provides functionalities for full image reading, delayed image reading, mosaic image reading, metadata reading, xarray coordinate plane attachment, cloud IO support, and saving to OME-TIFF. It also offers benchmarking and developer resources.
For similar jobs
cellseg_models.pytorch
cellseg-models.pytorch is a Python library built upon PyTorch for 2D cell/nuclei instance segmentation models. It provides multi-task encoder-decoder architectures and post-processing methods for segmenting cell/nuclei instances. The library offers high-level API to define segmentation models, open-source datasets for training, flexibility to modify model components, sliding window inference, multi-GPU inference, benchmarking utilities, regularization techniques, and example notebooks for training and finetuning models with different backbones.

aicsimageio
AICSImageIO is a Python tool for Image Reading, Metadata Conversion, and Image Writing for Microscopy Images. It supports various file formats like OME-TIFF, TIFF, ND2, DV, CZI, LIF, PNG, GIF, and Bio-Formats. Users can read and write metadata and imaging data, work with different file systems like local paths, HTTP URLs, s3fs, and gcsfs. The tool provides functionalities for full image reading, delayed image reading, mosaic image reading, metadata reading, xarray coordinate plane attachment, cloud IO support, and saving to OME-TIFF. It also offers benchmarking and developer resources.

KG_RAG
KG-RAG (Knowledge Graph-based Retrieval Augmented Generation) is a task agnostic framework that combines the explicit knowledge of a Knowledge Graph (KG) with the implicit knowledge of a Large Language Model (LLM). KG-RAG extracts "prompt-aware context" from a KG, which is defined as the minimal context sufficient enough to respond to the user prompt. This framework empowers a general-purpose LLM by incorporating an optimized domain-specific 'prompt-aware context' from a biomedical KG. KG-RAG is specifically designed for running prompts related to Diseases.

Scientific-LLM-Survey
Scientific Large Language Models (Sci-LLMs) is a repository that collects papers on scientific large language models, focusing on biology and chemistry domains. It includes textual, molecular, protein, and genomic languages, as well as multimodal language. The repository covers various large language models for tasks such as molecule property prediction, interaction prediction, protein sequence representation, protein sequence generation/design, DNA-protein interaction prediction, and RNA prediction. It also provides datasets and benchmarks for evaluating these models. The repository aims to facilitate research and development in the field of scientific language modeling.
biochatter
Generative AI models have shown tremendous usefulness in increasing accessibility and automation of a wide range of tasks. This repository contains the `biochatter` Python package, a generic backend library for the connection of biomedical applications to conversational AI. It aims to provide a common framework for deploying, testing, and evaluating diverse models and auxiliary technologies in the biomedical domain. BioChatter is part of the BioCypher ecosystem, connecting natively to BioCypher knowledge graphs.
ceLLama
ceLLama is a streamlined automation pipeline for cell type annotations using large-language models (LLMs). It operates locally to ensure privacy, provides comprehensive analysis by considering negative genes, offers efficient processing speed, and generates customized reports. Ideal for quick and preliminary cell type checks.
PINNACLE
PINNACLE is a flexible geometric deep learning approach that trains on contextualized protein interaction networks to generate context-aware protein representations. It provides protein representations split across various cell-type contexts from different tissues and organs. The tool can be fine-tuned to study the genomic effects of drugs and nominate promising protein targets and cell-type contexts for further investigation. PINNACLE exemplifies the paradigm of incorporating context-specific effects for studying biological systems, especially the impact of disease and therapeutics.
Taiyi-LLM
Taiyi (太一) is a bilingual large language model fine-tuned for diverse biomedical tasks. It aims to facilitate communication between healthcare professionals and patients, provide medical information, and assist in diagnosis, biomedical knowledge discovery, drug development, and personalized healthcare solutions. The model is based on the Qwen-7B-base model and has been fine-tuned using rich bilingual instruction data. It covers tasks such as question answering, biomedical dialogue, medical report generation, biomedical information extraction, machine translation, title generation, text classification, and text semantic similarity. The project also provides standardized data formats, model training details, model inference guidelines, and overall performance metrics across various BioNLP tasks.