
mercure
mercure DICOM Orchestrator
Stars: 64
mercure DICOM Orchestrator is a flexible solution for routing and processing DICOM files. It offers a user-friendly web interface and extensive monitoring functions. Custom processing modules can be implemented as Docker containers. Written in Python, it uses the DCMTK toolkit for DICOM communication. It can be deployed as a single-server installation using Docker Compose or as a scalable cluster installation using Nomad. mercure consists of service modules for receiving, routing, processing, dispatching, cleaning, web interface, and central monitoring.
README:
A flexible DICOM routing and processing solution with user-friendly web interface and extensive monitoring functions. Custom processing modules can be implemented as Docker containers. mercure has been written in the Python language and uses the DCMTK toolkit for the underlying DICOM communication. It can be deployed either as containerized single-server installation using Docker Compose, or as scalable cluster installation using Nomad. mercure consists of multiple service modules that handle different steps of the processing pipeline.
Installation instructions and usage information can be found in the project documentation:
https://mercure-imaging.org/docs/index.html
The receiver listens on a tcp port for incoming DICOM files. Received files are run through a preprocessing procedure, which extracts DICOM tag information and stores it in a json file.
The router module runs periodically and checks
- if the transfer of a DICOM series has finished (based on timeouts)
- if a routing rule triggers for the received series (or study)
If both conditions are met, the DICOM series (or study) is moved into a subdirectory of the outgoing folder or
processing folder (depending on the triggered rule), together with task file that describes the action to be performed.
If no rule applies, the DICOM series is placed in the discard folder.
The processor module runs periodically and checks for tasks submitted to the processing folder. It then locks the task and executes processing modules as defined in the task.json file. The requested processing module is started as Docker container, either on the same server or on a separate processing node (for Nomad installations). If results should be dispatched, the processed files are moved into a subfolder of the outgoing folder.
The dispatcher module runs periodically and checks
- if a transfer from the router or processor has finished
- if the series is not already being dispatched
- if at least one DICOM file is available
If the conditions are true, the information about the DICOM target node is read from the
task.json file and the images are sent to this node. After the transfer, the files
are moved to either the success or error folder.
The cleaner module runs periodically and checks
- if new series arrived in the
discardorsuccessfolder - if the move operation into these folder has finished
- if the predefined clean-up delay has elapsed (by default, 3 days)
If these conditions are true, series in the success and discard folders are deleted.
The webgui module provides a user-friendly web interface for configuring, controlling, and monitoring the server.
The bookkeeper module acts as central monitoring instance for all mercure services. The individual modules communicate with the bookkeeper via a TCP/IP connection. The submitted information is stored in a Postgres database.
For Tasks:
Click tags to check more tools for each tasksFor Jobs:
Alternative AI tools for mercure
Similar Open Source Tools

mercure
mercure DICOM Orchestrator is a flexible solution for routing and processing DICOM files. It offers a user-friendly web interface and extensive monitoring functions. Custom processing modules can be implemented as Docker containers. Written in Python, it uses the DCMTK toolkit for DICOM communication. It can be deployed as a single-server installation using Docker Compose or as a scalable cluster installation using Nomad. mercure consists of service modules for receiving, routing, processing, dispatching, cleaning, web interface, and central monitoring.

airbroke
Airbroke is an open-source error catcher tool designed for modern web applications. It provides a PostgreSQL-based backend with an Airbrake-compatible HTTP collector endpoint and a React-based frontend for error management. The tool focuses on simplicity, maintaining a small database footprint even under heavy data ingestion. Users can ask AI about issues, replay HTTP exceptions, and save/manage bookmarks for important occurrences. Airbroke supports multiple OAuth providers for secure user authentication and offers occurrence charts for better insights into error occurrences. The tool can be deployed in various ways, including building from source, using Docker images, deploying on Vercel, Render.com, Kubernetes with Helm, or Docker Compose. It requires Node.js, PostgreSQL, and specific system resources for deployment.
aigt
AIGT is a repository containing scripts for deep learning in guided medical interventions, focusing on ultrasound imaging. It provides a complete workflow from formatting and annotations to real-time model deployment. Users can set up an Anaconda environment, run Slicer notebooks, acquire tracked ultrasound data, and process exported data for training. The repository includes tools for segmentation, image export, and annotation creation.
cluster-toolkit
Cluster Toolkit is an open-source software by Google Cloud for deploying AI/ML and HPC environments on Google Cloud. It allows easy deployment following best practices, with high customization and extensibility. The toolkit includes tutorials, examples, and documentation for various modules designed for AI/ML and HPC use cases.
HuggingFists
HuggingFists is a low-code data flow tool that enables convenient use of LLM and HuggingFace models. It provides functionalities similar to Langchain, allowing users to design, debug, and manage data processing workflows, create and schedule workflow jobs, manage resources environment, and handle various data artifact resources. The tool also offers account management for users, allowing centralized management of data source accounts and API accounts. Users can access Hugging Face models through the Inference API or locally deployed models, as well as datasets on Hugging Face. HuggingFists supports breakpoint debugging, branch selection, function calls, workflow variables, and more to assist users in developing complex data processing workflows.
nx_open
The `nx_open` repository contains open-source components for the Network Optix Meta Platform, used to build products like Nx Witness Video Management System. It includes source code, specifications, and a Desktop Client. The repository is licensed under Mozilla Public License 2.0. Users can build the Desktop Client and customize it using a zip file. The build environment supports Windows, Linux, and macOS platforms with specific prerequisites. The repository provides scripts for building, signing executable files, and running the Desktop Client. Compatibility with VMS Server versions is crucial, and automatic VMS updates are disabled for the open-source Desktop Client.
dlio_benchmark
DLIO is an I/O benchmark tool designed for Deep Learning applications. It emulates modern deep learning applications using Benchmark Runner, Data Generator, Format Handler, and I/O Profiler modules. Users can configure various I/O patterns, data loaders, data formats, datasets, and parameters. The tool is aimed at emulating the I/O behavior of deep learning applications and provides a modular design for flexibility and customization.
AirSane
AirSane is a SANE frontend and scanner server that supports Apple's AirScan protocol. It automatically detects scanners and publishes them through mDNS. Acquired images can be transferred in JPEG, PNG, and PDF/raster format. The tool is intended to be used with AirScan/eSCL clients such as Apple's Image Capture, sane-airscan on Linux, and the eSCL client built into Windows 10 and 11. It provides a simple web interface and encodes images on-the-fly to keep memory/storage demands low, making it suitable for devices like Raspberry Pi. Authentication and secure communication are supported in conjunction with a proxy server like nginx. AirSane has been reverse-engineered from Apple's AirScanScanner client communication protocol and offers a range of installation and configuration options for different operating systems.
serverless-pdf-chat
The serverless-pdf-chat repository contains a sample application that allows users to ask natural language questions of any PDF document they upload. It leverages serverless services like Amazon Bedrock, AWS Lambda, and Amazon DynamoDB to provide text generation and analysis capabilities. The application architecture involves uploading a PDF document to an S3 bucket, extracting metadata, converting text to vectors, and using a LangChain to search for information related to user prompts. The application is not intended for production use and serves as a demonstration and educational tool.
minio
MinIO is a High Performance Object Storage released under GNU Affero General Public License v3.0. It is API compatible with Amazon S3 cloud storage service. Use MinIO to build high performance infrastructure for machine learning, analytics and application data workloads.
holoscan-sdk
The Holoscan SDK is part of NVIDIA Holoscan, the AI sensor processing platform that combines hardware systems for low-latency sensor and network connectivity, optimized libraries for data processing and AI, and core microservices to run streaming, imaging, and other applications, from embedded to edge to cloud. It can be used to build streaming AI pipelines for a variety of domains, including Medical Devices, High Performance Computing at the Edge, Industrial Inspection and more.
aws-lex-web-ui
The AWS Lex Web UI is a sample Amazon Lex web interface that provides a chatbot UI component for integration into websites. It supports voice and text interactions, Lex response cards, and programmable configuration using JavaScript. The interface can be used as a full-page chatbot UI or embedded as a widget. It offers mobile-ready responsive UI, seamless voice-text switching, and interactive messaging support. The project includes CloudFormation templates for easy deployment and customization. Users can modify configurations, integrate the UI into existing sites, and deploy using various methods like CloudFormation, pre-built libraries, or npm installation.
serena
Serena is a powerful coding agent that integrates with existing LLMs to provide essential semantic code retrieval and editing tools. It is free to use and does not require API keys or subscriptions. Serena can be used for coding tasks such as analyzing, planning, and editing code directly on your codebase. It supports various programming languages and offers semantic code analysis capabilities through language servers. Serena can be integrated with different LLMs using the model context protocol (MCP) or Agno framework. The tool provides a range of functionalities for code retrieval, editing, and execution, making it a versatile coding assistant for developers.
aici
The Artificial Intelligence Controller Interface (AICI) lets you build Controllers that constrain and direct output of a Large Language Model (LLM) in real time. Controllers are flexible programs capable of implementing constrained decoding, dynamic editing of prompts and generated text, and coordinating execution across multiple, parallel generations. Controllers incorporate custom logic during the token-by-token decoding and maintain state during an LLM request. This allows diverse Controller strategies, from programmatic or query-based decoding to multi-agent conversations to execute efficiently in tight integration with the LLM itself.
trinityX
TrinityX is an open-source HPC, AI, and cloud platform designed to provide all services required in a modern system, with full customization options. It includes default services like Luna node provisioner, OpenLDAP, SLURM or OpenPBS, Prometheus, Grafana, OpenOndemand, and more. TrinityX also sets up NFS-shared directories, OpenHPC applications, environment modules, HA, and more. Users can install TrinityX on Enterprise Linux, configure network interfaces, set up passwordless authentication, and customize the installation using Ansible playbooks. The platform supports HA, OpenHPC integration, and provides detailed documentation for users to contribute to the project.
aide
AIDE (Advanced Intrusion Detection Environment) is a tool for monitoring file system changes. It can be used to detect unauthorized changes to monitored files and directories. AIDE was written to be a simple and free alternative to Tripwire. Features currently included in AIDE are as follows: o File attributes monitored: permissions, inode, user, group file size, mtime, atime, ctime, links and growing size. o Checksums and hashes supported: SHA1, MD5, RMD160, and TIGER. CRC32, HAVAL and GOST if Mhash support is compiled in. o Plain text configuration files and database for simplicity. o Rules, variables and macros that can be customized to local site or system policies. o Powerful regular expression support to selectively include or exclude files and directories to be monitored. o gzip database compression if zlib support is compiled in. o Free software licensed under the GNU General Public License v2.
For similar tasks

mercure
mercure DICOM Orchestrator is a flexible solution for routing and processing DICOM files. It offers a user-friendly web interface and extensive monitoring functions. Custom processing modules can be implemented as Docker containers. Written in Python, it uses the DCMTK toolkit for DICOM communication. It can be deployed as a single-server installation using Docker Compose or as a scalable cluster installation using Nomad. mercure consists of service modules for receiving, routing, processing, dispatching, cleaning, web interface, and central monitoring.
For similar jobs
Generative-AI-Drug-Discovery
Generative-AI-Drug-Discovery is a public repository on GitHub focused on using tensor network machine learning approaches to accelerate GenAI for drug discovery. The repository aims to implement effective architectures and methodologies into Large Language Models (LLMs) to enhance Drug Discovery Generative AI performance.
AI2BMD
AI2BMD is a program for efficiently simulating protein molecular dynamics with ab initio accuracy. The repository contains datasets, simulation programs, and public materials related to AI2BMD. It provides a Docker image for easy deployment and a standalone launcher program. Users can run simulations by downloading the launcher script and specifying simulation parameters. The repository also includes ready-to-use protein structures for testing. AI2BMD is designed for x86-64 GNU/Linux systems with recommended hardware specifications. The related research includes model architectures like ViSNet, Geoformer, and fine-grained force metrics for MLFF. Citation information and contact details for the AI2BMD Team are provided.

mercure
mercure DICOM Orchestrator is a flexible solution for routing and processing DICOM files. It offers a user-friendly web interface and extensive monitoring functions. Custom processing modules can be implemented as Docker containers. Written in Python, it uses the DCMTK toolkit for DICOM communication. It can be deployed as a single-server installation using Docker Compose or as a scalable cluster installation using Nomad. mercure consists of service modules for receiving, routing, processing, dispatching, cleaning, web interface, and central monitoring.
ProLLM
ProLLM is a framework that leverages Large Language Models to interpret and analyze protein sequences and interactions through natural language processing. It introduces the Protein Chain of Thought (ProCoT) method to transform complex protein interaction data into intuitive prompts, enhancing predictive accuracy by incorporating protein-specific embeddings and fine-tuning on domain-specific datasets.
fuse-med-ml
FuseMedML is a Python framework designed to accelerate machine learning-based discovery in the medical field by promoting code reuse. It provides a flexible design concept where data is stored in a nested dictionary, allowing easy handling of multi-modality information. The framework includes components for creating custom models, loss functions, metrics, and data processing operators. Additionally, FuseMedML offers 'batteries included' key components such as fuse.data for data processing, fuse.eval for model evaluation, and fuse.dl for reusable deep learning components. It supports PyTorch and PyTorch Lightning libraries and encourages the creation of domain extensions for specific medical domains.
MedLLMsPracticalGuide
This repository serves as a practical guide for Medical Large Language Models (Medical LLMs) and provides resources, surveys, and tools for building, fine-tuning, and utilizing LLMs in the medical domain. It covers a wide range of topics including pre-training, fine-tuning, downstream biomedical tasks, clinical applications, challenges, future directions, and more. The repository aims to provide insights into the opportunities and challenges of LLMs in medicine and serve as a practical resource for constructing effective medical LLMs.
hi-ml
The Microsoft Health Intelligence Machine Learning Toolbox is a repository that provides low-level and high-level building blocks for Machine Learning / AI researchers and practitioners. It simplifies and streamlines work on deep learning models for healthcare and life sciences by offering tested components such as data loaders, pre-processing tools, deep learning models, and cloud integration utilities. The repository includes two Python packages, 'hi-ml-azure' for helper functions in AzureML, 'hi-ml' for ML components, and 'hi-ml-cpath' for models and workflows related to histopathology images.
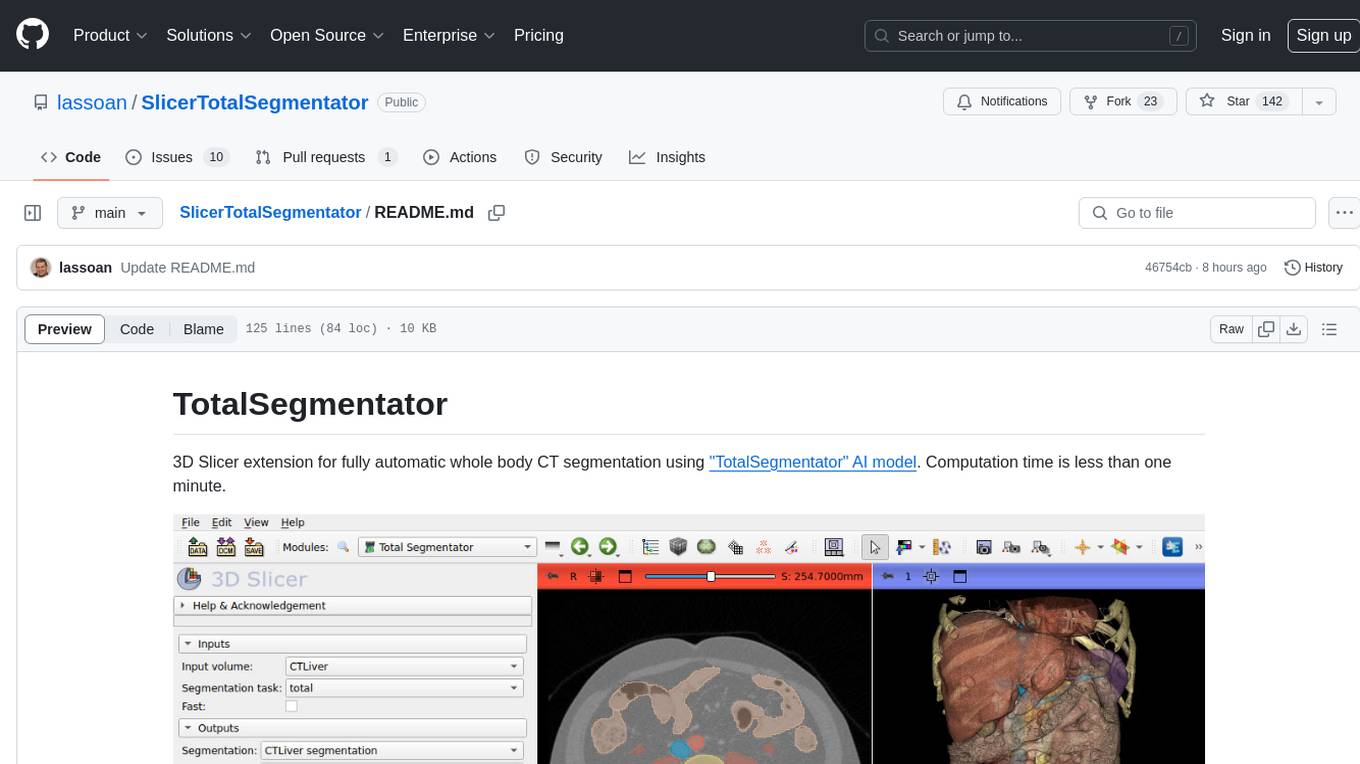
SlicerTotalSegmentator
TotalSegmentator is a 3D Slicer extension designed for fully automatic whole body CT segmentation using the 'TotalSegmentator' AI model. The computation time is less than one minute, making it efficient for research purposes. Users can set up GPU acceleration for faster segmentation. The tool provides a user-friendly interface for loading CT images, creating segmentations, and displaying results in 3D. Troubleshooting steps are available for common issues such as failed computation, GPU errors, and inaccurate segmentations. Contributions to the extension are welcome, following 3D Slicer contribution guidelines.


