
MedLLMsPracticalGuide
A curated list of practical guide resources of Medical LLMs (Medical LLMs Tree, Tables, and Papers)
Stars: 1310
This repository serves as a practical guide for Medical Large Language Models (Medical LLMs) and provides resources, surveys, and tools for building, fine-tuning, and utilizing LLMs in the medical domain. It covers a wide range of topics including pre-training, fine-tuning, downstream biomedical tasks, clinical applications, challenges, future directions, and more. The repository aims to provide insights into the opportunities and challenges of LLMs in medicine and serve as a practical resource for constructing effective medical LLMs.
README:
This is an actively updated list of practical guide resources for Medical Large Language Models (Medical LLMs). It's based on our survey paper:
[Nature Reviews Bioengineering] A Survey of Large Language Models in Medicine: Progress, Application, and Challenge
Hongjian Zhou1,*, Fenglin Liu1,*, Boyang Gu2,*, Xinyu Zou3,*, Jinfa Huang4,*, Jinge Wu5, Yiru Li6, Sam S. Chen7, Peilin Zhou8, Junling Liu9, Yining Hua10, Chengfeng Mao11, Chenyu You12, Xian Wu13, Yefeng Zheng13, Lei Clifton1, Zheng Li14,†, Jiebo Luo4,†, David A. Clifton1,†. (*Core Contributors, †Corresponding Authors)
1University of Oxford, 2Imperial College London, 3University of Waterloo, 4University of Rochester, 5University College London, 6Western University, 7University of Georgia, 8Hong Kong University of Science and Technology (Guangzhou), 9Alibaba, 10Harvard T.H. Chan School of Public Health, 11MIT, 12Yale University, 13Tencent, 14Amazon
[2025-01-20] 🎉🎉🎉 Big News! Our paper has been accepted by Nature Reviews Bioengineering and GitHub Repo has reached 1,300 🌟!
[2023-11-09] We have released the repository and survey.
If you want to add your work or model to this list, please do not hesitate to email [email protected] and [email protected] or pull requests. Markdown format:
* [**Name of Conference or Journal + Year**] Paper Name. [[paper]](link) [[code]](link)Goal 1: Surpassing Human-Level Expertise.
Goal 2: Emergent Properties of Medical LLM with the Model Size Scaling Up.
This survey provides a comprehensive overview of the principles, applications, and challenges faced by LLMs in medicine. We address the following specific questions:
- How should medical LLMs be built?
- What are the measures for the downstream performance of medical LLMs?
- How should medical LLMs be utilized in real-world clinical practice?
- What challenges arise from the use of medical LLMs?
- How should we better construct and utilize medical LLMs?
This survey aims to provide insights into the opportunities and challenges of LLMs in medicine, and serve as a practical resource for constructing effective medical LLMs.
- 📣 Update News
- ⚡ Contributing
- 🤔 What are the Goals of the Medical LLM?
- 🤗 What is This Survey About?
- Table of Contents
- 🔥 Practical Guide for Building Pipeline
- 📊 Practical Guide for Medical Data
- 🗂️ Downstream Biomedical Tasks
- ✨ Practical Guide for Clinical Applications
- ⚔️ Practical Guide for Challenges
- 🚀 Practical Guide for Future Directions
- 👍 Acknowledgement
- 📑 Citation
♥️ Contributors
- [Nature Medicine, 2024] BiomedGPT A generalist vision–language foundation model for diverse biomedical tasks paper
- [Nature, 2023] NYUTron Health system-scale language models are all-purpose prediction engines paper
- [Arxiv, 2023] OphGLM: Training an Ophthalmology Large Language-and-Vision Assistant based on Instructions and Dialogue. paper
- [npj Digital Medicine, 2023] GatorTronGPT: A Study of Generative Large Language Model for Medical Research and Healthcare. paper
- [Bioinformatics, 2023] MedCPT: Contrastive Pre-trained Transformers with Large-scale Pubmed Search Logs for Zero-shot Biomedical Information Retrieval. paper
- [Bioinformatics, 2022] BioGPT: Generative Pre-trained Transformer for Biomedical Text Generation and Mining. paper
- [NeurIPS, 2022] DRAGON: Deep Bidirectional Language-Knowledge Graph Pretraining. paper code
- [ACL, 2022] BioLinkBERT/LinkBERT: Pretraining Language Models with Document Links. paper code
- [npj Digital Medicine, 2022] GatorTron: A Large Language Model for Electronic Health Records. paper
- [HEALTH, 2021] PubMedBERT: Domain-specific Language Model Pretraining for Biomedical Natural Language Processing. paper
- [Bioinformatics, 2020] BioBERT: A Pre-trained Biomedical Language Representation Model for Biomedical Text Mining. paper
- [ENNLP, 2019] SciBERT: A Pretrained Language Model for Scientific Text. paper
- [NAACL Workshop, 2019] ClinicalBERT: Publicly Available Clinical BERT Embeddings. paper
- [BioNLP Workshop, 2019] BlueBERT: Transfer Learning in Biomedical Natural Language Processing: An Evaluation of BERT and ELMo on Ten Benchmarking Datasets. paper
- [Nature Communications, 2024.9] MMed-Llama3: Towards building multilingual language model for medicine. [paper] [code]
- [Arxiv, 2024.8] Med42-v2: A Suite of Clinical LLMs. paper Model
- [JAMIA, 2024.5] Internist.ai 7b Impact of high-quality, mixed-domain data on the performance of medical language models paper Model
- [Huggingface, 2024.5] OpenBioLLM-70b: Advancing Open-source Large Language Models in Medical Domain Model
- [Huggingface, 2024.5] MedLllama3 model
- [Arxiv, 2024.5] Aloe: A Family of Fine-tuned Open Healthcare LLMs. paper Model
- [Arxiv, 2024.4] Med-Gemini Capabilities of Gemini Models in Medicine. paper
- [Arxiv, 2024.2] BioMistral A Collection of Open-Source Pretrained Large Language Models for Medical Domains. paper
- [Arxiv, 2023.12] From Beginner to Expert: Modeling Medical Knowledge into General LLMs. paper
- [Arxiv, 2023.11] Taiyi: A Bilingual Fine-Tuned Large Language Model for Diverse Biomedical Tasks. paper code
- [Arxiv, 2023.10] AlpaCare: Instruction-tuned Large Language Models for Medical Application. paper code
- [Arxiv, 2023.10] BianQue: Balancing the Questioning and Suggestion Ability of Health LLMs with Multi-turn Health Conversations Polished by ChatGPT. paper
- [Arxiv, 2023.10] Qilin-Med: Multi-stage Knowledge Injection Advanced Medical Large Language Model. paper
- [Arxiv, 2023.10] Qilin-Med-VL: Towards Chinese Large Vision-Language Model for General Healthcare. paper
- [Arxiv, 2023.10] MEDITRON-70B: Scaling Medical Pretraining for Large Language Models. paper
- [AAAI, 2024/2023.10] Med42: Evaluating Fine-Tuning Strategies for Medical LLMs: Full-Parameter vs. Parameter-Efficient Approaches. paper Model
- [Arxiv, 2023.9] CPLLM: Clinical Prediction with Large Language Models. paper
- [Arxiv, 2023.8] BioMedGPT/OpenBioMed Open Multimodal Generative Pre-trained Transformer for BioMedicine. paper code
- [Nature Digital Medicine, 2023.8] Large Language Models to Identify Social Determinants of Health in Electronic Health Records. paper [code]
- [Arxiv, 2023.8] Zhongjing: Enhancing the Chinese medical capabilities of large language model through expert feedback and real-world multi-turn dialogue. paper
- [Arxiv, 2023.7] Med-Flamingo: Med-Flamingo: a Multimodal Medical Few-shot Learner. paper code
- [Arxiv, 2023.6] ClinicalGPT: Large Language Models Finetuned with Diverse Medical Data and Comprehensive Evaluation. 2023. paper
- [Cureus, 2023.6] ChatDoctor: A Medical Chat Model Fine-Tuned on a Large Language Model Meta-AI (LLaMA) Using Medical Domain Knowledge. paper
- [NeurIPS Datasets/Benchmarks Track, 2023.6] LLaVA-Med: Training a large language-and-vision assistant for biomedicine in one day. paper
- [Arxiv, 2023.6] MedPaLM 2: Towards expert-level medical question answering with large language models. paper
- [Arxiv, 2023.5] Clinical Camel: An Open-Source Expert-Level Medical Language Model with Dialogue-Based Knowledge Encoding. paper
- [Arxiv, 2023.5] BiomedGPT: A Generalist Vision-Language Foundation Model for Diverse Biomedical Tasks. paper
- [Arxiv, 2023.5] HuatuoGPT: HuatuoGPT, towards Taming Language Model to Be a Doctor. paper
- [Arxiv, 2023.4] Baize-healthcare: An open-source chat model with parameter-efficient tuning on self-chat data. paper
- [Arxiv, 2023.4] Visual Med-Alpeca: A parameter-efficient biomedical llm with visual capabilities. github
- [Arxiv, 2023.4] PMC-LLaMA: Further finetuning llama on medical papers. paper
- [Arxiv, 2023.4] MedPaLM M: Towards Generalist Biomedical AI. paper code
- [Arxiv, 2023.4] BenTsao/Huatuo: Tuning llama model with chinese medical knowledge. paper
- [Github, 2023.4] ChatGLM-Med: ChatGLM-Med: 基于中文医学知识的ChatGLM模型微调. github
- [Arxiv, 2023.4] DoctorGLM: Fine-tuning your chinese doctor is not a herculean task. paper
- [NEJM AI, 2024] GPT-4 for Information Retrieval and Comparison of Medical Oncology Guidelines. paper
- [Arxiv, 2023.11] MedPrompt: Can Generalist Foundation Models Outcompete Special-Purpose Tuning? Case Study in Medicine. paper
- [Arxiv, 2023.8] Dr. Knows: Leveraging a medical knowledge graph into large language models for diagnosis prediction. paper
- [Arxiv, 2023.3] DelD-GPT: Zero-shot medical text de-identification by gpt-4. paper code
- [Arxiv, 2023.2/5] ChatCAD/ChatCAD+: Interactive computer-aided diagnosis on medical image using large language models. paper code
- [Nature, 2022.12] MedPaLM: Large language models encode clinical knowledge. paper
- [Arxiv, 2022.7/2023.12] Can large language models reason about medical questions? paper
- Drugs.com
- DrugBank
- NHS Health
- NHS Medicine
- Unified Medical Language System (UMLS)
- The Human Phenotype Ontology
- Center for Disease Control and Prevention
- National Institute for Health and Care Excellence
- World Health Organization
- [NEJM AI, 2024] Clinical Text Datasets for Medical Artificial Intelligence and Large Language Models — A Systematic Review paper
- [npj Digital Medicine, 2023] EHRs: A Study of Generative Large Language Model for Medical Research and Healthcare. paper
- [Arxiv, 2023] Guidelines: A high-quality collection of clinical practice guidelines (CPGs) for the medical training of LLMs. dataset
- [Arxiv, 2023] GAP-REPLAY: Scaling Medical Pretraining for Large Language Models. paper
- [npj Digital Medicine, 2022] EHRs: A large language model for electronic health records. paper
- [National Library of Medicine, 2022] PubMed: National Institutes of Health. PubMed Data. database
- [Arxiv, 2020] PubMed: The pile: An 800gb dataset of diverse text for language modeling. paper code
- [EMNLP, 2020] MedDialog: Meddialog: Two large-scale medical dialogue datasets. paper code
- [NAACL, 2018] Literature: Construction of the literature graph in semantic scholar. paper
- [Scientific Data, 2016] MIMIC-III: MIMIC-III, a freely accessible critical care database. paper
- MMedC: Towards building multilingual language model for medicine. [paper] [code] [huggingface]
- MedTrinity-25M: A Large-scale Multimodal Dataset with Multigranular Annotations for Medicine. 2024. github paper
- cMeKG: Chinese Medical Knowledge Graph. 2023. github
- CMD.: Chinese medical dialogue data. 2023. repo
- BianQueCorpus: BianQue: Balancing the Questioning and Suggestion Ability of Health LLMs with Multi-turn Health Conversations Polished by ChatGPT. 2023. paper
- MD-EHR: ClinicalGPT: Large Language Models Finetuned with Diverse Medical Data and Comprehensive Evaluation. 2023. paper
- VariousMedQA: Multi-scale attentive interaction networks for chinese medical question answer selection. 2018. paper
- VariousMedQA: What disease does this patient have? a large-scale open domain question answering dataset from medical exams. 2021. paper
- MedDialog: Meddialog: Two large-scale medical dialogue datasets. 2020. paper
- ChiMed: Qilin-Med: Multi-stage Knowledge Injection Advanced Medical Large Language Model. 2023. paper
- ChiMed-VL: Qilin-Med-VL: Towards Chinese Large Vision-Language Model for General Healthcare. 2023. paper
- Healthcare Magic: Healthcare Magic. link,huggingface
- ICliniq: ICliniq. platform
- Hybrid SFT: HuatuoGPT, towards Taming Language Model to Be a Doctor. 2023. paper
- PMC-15M: Large-scale domain-specific pretraining for biomedical vision-language processing. 2023. paper
- MedQuAD: A question-entailment approach to question answering. 2019. paper
- VariousMedQA: Visual med-alpaca: A parameter-efficient biomedical llm with visual capabilities. 2023. repo
- CMtMedQA:Zhongjing: Enhancing the Chinese medical capabilities of large language model through expert feedback and real-world multi-turn dialogue. 2023. paper
- MTB: Med-flamingo: a multimodal medical few-shot learner. 2023. paper
- PMC-OA: Pmc-clip: Contrastive language-image pre-training using biomedical documents. 2023. paper
- Medical Meadow: MedAlpaca--An Open-Source Collection of Medical Conversational AI Models and Training Data. 2023. paper
- Literature: S2ORC: The semantic scholar open research corpus. 2019. paper
- MedC-I: Pmc-llama: Further finetuning llama on medical papers. 2023. paper
- ShareGPT: Sharegpt. 2023. platform
- PubMed: National Institutes of Health. PubMed Data. In National Library of Medicine. 2022. database
- MedQA: What disease does this patient have? a large-scale open domain question answering dataset from medical exams. 2021. paper
- MultiMedQA: Towards expert-level medical question answering with large language models. 2023. paper
- MultiMedBench: Towards generalist biomedical ai. 2023. paper
- MedInstruct-52: Instruction-tuned Large Language Models for Medical Application. 2023. paper
- eICU-CRD: The eicu collaborative research database, a freely available multi-center database for critical care research. 2018. paper
- MIMIC-IV: MIMIC-IV, a freely accessible electronic health record dataset. 2023. paper database
- PMC-Patients: 167k open patient summaries. 2023. paper database
- Open Medical-LLM Leaderboard: MedQA (USMLE), PubMedQA, MedMCQA, and subsets of MMLU related to medicine and biology. Leaderboard
- ReXrank: A Public Leaderboard for AI-Powered Radiology Report Generation paper [[paper]] [[code]] (https://github.com/rajpurkarlab/ReXrank)
- PubMed: National Institutes of Health. PubMed Data. In National Library of Medicine. database
- PMC: National Institutes of Health. PubMed Central Data. In National Library of Medicine. database
- CORD-19: Cord-19: The covid-19 open research dataset 2020. paper
- MentSum: Mentsum: A resource for exploring summarization of mental health online posts 2022. paper
- MeQSum: On the summarization of consumer health questions 2019. paper
- MedQSum: Enhancing Large Language Models’ Utility for Medical Question-Answering: A Patient Health Question Summarization Approach. [paper] [code]
- MultiCochrane: Multilingual Simplification of Medical Texts 2023. paper
- AutoMeTS: AutoMeTS: the autocomplete for medical text simplification 2020. paper
- CareQA: CareQA: A multichoice question answering dataset based on the access exam for Spanish Specialised Healthcare Training (FSE).paper dataset
- BioASQ-QA: BioASQ-QA: A manually curated corpus for Biomedical Question Answering 2023. paper
- emrQA: emrqa: A large corpus for question answering on electronic medical records 2018. paper
- CliCR: CliCR: a dataset of clinical case reports for machine reading comprehension 2018. paper
- PubMedQA: Pubmedqa: A dataset for biomedical research question answering 2019. paper
- COVID-QA: COVID-QA: A question answering dataset for COVID-19 2020. paper
- MASH-QA: Question answering with long multiple-span answers 2020. paper
- Health-QA: A hierarchical attention retrieval model for healthcare question answering 2019. paper
- MedQA: What disease does this patient have? a large-scale open domain question answering dataset from medical exams 2021. paper
- MedMCQA: Medmcqa: A large-scale multi-subject multi-choice dataset for medical domain question answering 2022. paper
- MMLU (Clinical Knowledge): Measuring massive multitask language understanding 2020. paper
- MMLU (College Medicine): Measuring massive multitask language understanding 2020. paper
- MMLU (Professional Medicine): Measuring massive multitask language understanding 2020. paper
- [Arxiv 2024] MediQ: Question-Asking LLMs for Adaptive and Reliable Clinical Reasoning. [paper] [code].
- [Arxiv, 2024.10] Named Clinical Entity Recognition Benchmark paper Leaderboard
- NCBI Disease: NCBI disease corpus: a resource for disease name recognition and concept normalization 2014. paper
- JNLPBA: Introduction to the bio-entity recognition task at JNLPBA 2004. paper
- GENIA: GENIA corpus--a semantically annotated corpus for bio-textmining 2003. paper
- BC5CDR: BioCreative V CDR task corpus: a resource for chemical disease relation extraction 2016. paper
- BC4CHEMD: The CHEMDNER corpus of chemicals and drugs and its annotation principles 2015. paper
- BioRED: BioRED: a rich biomedical relation extraction dataset 2022. paper
- CMeEE: Cblue: A chinese biomedical language understanding evaluation benchmark 2021. paper
- NLM-Chem-BC7: NLM-Chem-BC7: manually annotated full-text resources for chemical entity annotation and indexing in biomedical articles 2022. paper
- ADE: Development of a benchmark corpus to support the automatic extraction of drug-related adverse effects from medical case reports 2012. paper
- 2012 i2b2: Evaluating temporal relations in clinical text: 2012 i2b2 challenge 2013. paper
- 2014 i2b2/UTHealth (Track 1): Annotating longitudinal clinical narratives for de-identification: The 2014 i2b2/UTHealth corpus 2015. paper
- 2018 n2c2 (Track 2): 2018 n2c2 shared task on adverse drug events and medication extraction in electronic health records 2020. paper
- Cadec: Cadec: A corpus of adverse drug event annotations 2015. paper
- DDI: Semeval-2013 task 9: Extraction of drug-drug interactions from biomedical texts (ddiextraction 2013) 2013. paper
- PGR: A silver standard corpus of human phenotype-gene relations 2019. paper
- EU-ADR: The EU-ADR corpus: annotated drugs, diseases, targets, and their relationships 2012. paper
- [BioCreative VII Challenge, 2021] Medications detection in tweets using transformer networks and multi-task learning. [paper] [code]
- BC5CDR: BioCreative V CDR task corpus: a resource for chemical disease relation extraction 2016. paper
- BioRED: BioRED: a rich biomedical relation extraction dataset 2022. paper
- ADE: Development of a benchmark corpus to support the automatic extraction of drug-related adverse effects from medical case reports 2012. paper
- 2018 n2c2 (Track 2): 2018 n2c2 shared task on adverse drug events and medication extraction in electronic health records 2020. paper
- 2010 i2b2/VA: 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text 2011. paper
- ChemProt: Overview of the BioCreative VI chemical-protein interaction Track 2017. database
- GDA: Renet: A deep learning approach for extracting gene-disease associations from literature 2019. paper
- DDI: Semeval-2013 task 9: Extraction of drug-drug interactions from biomedical texts (ddiextraction 2013) 2013. paper
- GAD: The genetic association database 2004. paper
- 2012 i2b2: Evaluating temporal relations in clinical text: 2012 i2b2 challenge 2013. paper
- PGR: A silver standard corpus of human phenotype-gene relations 2019. paper
- EU-ADR: The EU-ADR corpus: annotated drugs, diseases, targets, and their relationships 2012. paper
- OpiateID: Identifying Self-Disclosures of Use, Misuse and Addiction in Community-based Social Media Posts. paper code
- ADE: Development of a benchmark corpus to support the automatic extraction of drug-related adverse effects from medical case reports 2012. paper
- 2014 i2b2/UTHealth (Track 2): Annotating longitudinal clinical narratives for de-identification: The 2014 i2b2/UTHealth corpus 2015. paper
- HoC: Automatic semantic classification of scientific literature according to the hallmarks of cancer 2016. paper
- OHSUMED: OHSUMED: An interactive retrieval evaluation and new large test collection for research 1994. paper
- WNUT-2020 Task 2: WNUT-2020 task 2: identification of informative COVID-19 english tweets 2020. paper
- Medical Abstracts: Evaluating unsupervised text classification: zero-shot and similarity-based approaches 2022. paper
- MIMIC-III: MIMIC-III, a freely accessible critical care database 2016. paper
- MedNLI: Lessons from natural language inference in the clinical domain 2018. paper
- BioNLI: BioNLI: Generating a Biomedical NLI Dataset Using Lexico-semantic Constraints for Adversarial Examples 2022. paper
- MedSTS: MedSTS: a resource for clinical semantic textual similarity 2020. paper
- 2019 n2c2/OHNLP: The 2019 n2c2/ohnlp track on clinical semantic textual similarity: overview 2020. paper
- BIOSSES: BIOSSES: a semantic sentence similarity estimation system for the biomedical domain 2017. paper
- TREC-COVID: TREC-COVID: constructing a pandemic information retrieval test collection 2021. paper
- NFCorpus: A full-text learning to rank dataset for medical information retrieval 2016. paper
- BioASQ (BEIR): A heterogenous benchmark for zero-shot evaluation of information retrieval models 2021. paper
- [Arxiv, 2024] Medical Graph RAG: Towards Safe Medical Large Language Model via Graph Retrieval-Augmented Generation. paper
- [NEJM AI, 2024] GPT-4 for Information Retrieval and Comparison of Medical Oncology Guidelines. paper
- [Arxiv, 2023] Think and Retrieval: A Hypothesis Knowledge Graph Enhanced Medical Large Language Models. paper
- [JASN, 2023] Retrieve, Summarize, and Verify: How Will ChatGPT Affect Information Seeking from the Medical Literature? paper
- [NAACL Findings, 2024] Identifying Self-Disclosures of Use, Misuse and Addiction in Community-based Social Media Posts. paper code
- [Nature, 2023] NYUTron Health system-scale language models are all-purpose prediction engines paper
- [Arxiv, 2023] Leveraging a medical knowledge graph into large language models for diagnosis prediction. paper
- [Arxiv, 2023] ChatCAD+/Chatcad: Interactive computer-aided diagnosis on medical image using large language models. paper code
- [Cancer Inform, 2023] Designing a Deep Learning-Driven Resource-Efficient Diagnostic System for Metastatic Breast Cancer: Reducing Long Delays of Clinical Diagnosis and Improving Patient Survival in Developing Countries. paper
- [Nature Medicine, 2023] Large language models in medicine. paper
- [Nature Medicine, 2022] AI in health and medicine. paper
- [NEJM AI, 2024] Large Language Models Are Poor Medical Coders — Benchmarking of Medical Code Querying. paper
- [JMAI, 2023] Applying large language model artificial intelligence for retina International Classification of Diseases (ICD) coding. paper
- [ClinicalNLP Workshop, 2022] PLM-ICD: automatic ICD coding with pretrained language models. paper code
- [Nature Medicine, 2024] Adapted large language models can outperform medical experts in clinical text summarization. paper
- [Arxiv, 2023] Can GPT-4V (ision) Serve Medical Applications? Case Studies on GPT-4V for Multimodal Medical Diagnosis. paper
- [Arxiv, 2023] Qilin-Med-VL: Towards Chinese Large Vision-Language Model for General Healthcare. paper
- [Arxiv, 2023] Customizing General-Purpose Foundation Models for Medical Report Generation. paper
- [Arxiv, 2023] Towards generalist foundation model for radiology. paper code
- [Arxiv, 2023] Clinical Text Summarization: Adapting Large Language Models Can Outperform Human Experts. 2023. paper project code
- [Arxiv, 2023] MAIRA-1: A specialised large multimodal model for radiology report generation. paper project
- [Arxiv, 2023] Consensus, dissensus and synergy between clinicians and specialist foundation models in radiology report generation. paper
- [Lancet Digit Health, 2023] Using ChatGPT to write patient clinic letters. paper
- [Lancet Digit Health, 2023] ChatGPT: the future of discharge summaries?. paper
- [Arxiv, 2023.2/5] ChatCAD/ChatCAD+: Interactive computer-aided diagnosis on medical image using large language models. paper code
- [JMIR, 2023] Large Language Models in Medical Education: Opportunities, Challenges, and Future Directions. paper
- [JMIR, 2023] The Advent of Generative Language Models in Medical Education. paper
- [Korean J Med Educ., 2023] The impending impacts of large language models on medical education. paper
- [Healthcare, 2023]Leveraging Generative AI and Large Language Models: A Comprehensive Roadmap for Healthcare Integration. paper
- [ICARM, 2023] A Nested U-Structure for Instrument Segmentation in Robotic Surgery. paper
- [Appl. Sci., 2023] The multi-trip autonomous mobile robot scheduling problem with time windows in a stochastic environment at smart hospitals. paper
- [Arxiv, 2023] GRID: Scene-Graph-based Instruction-driven Robotic Task Planning. paper
- [I3CE, 2023] Trust in Construction AI-Powered Collaborative Robots: A Qualitative Empirical Analysis. paper
- [STAR, 2016] Advanced robotics for medical rehabilitation. paper
- [New Biotechnology, 2023] Machine translation of standardised medical terminology using natural language processing: A Scoping Review. paper
- [JMIR, 2023] The Advent of Generative Language Models in Medical Education. paper
- [Arxiv, 2024] Large Language Models in Mental Health Care: a Scoping Review. paper
- [Arxiv, 2023] PsyChat: A Client-Centric Dialogue System for Mental Health Support. paper code
- [Arxiv, 2023] Benefits and Harms of Large Language Models in Digital Mental Health. paper
- [CIKM, 2023] ChatCounselor: A Large Language Models for Mental Health Support. paper code
- [HCII, 2023] Tell me, what are you most afraid of? Exploring the Effects of Agent Representation on Information Disclosure in Human-Chatbot Interaction. paper
- [IJSR, 2023] A Brief Wellbeing Training Session Delivered by a Humanoid Social Robot: A Pilot Randomized Controlled Trial. paper
- [CHB, 2015] Real conversations with artificial intelligence: A comparison between human–human online conversations and human–chatbot conversations. paper
- [Arxiv, 2024] Chain-of-verification reduces hallucination in large language models. paper
- [ACM Computing Surveys, 2023] Survey of hallucination in natural language generation. paper
- [EMNLP, 2023] Med-halt: Medical domain hallucination test for large language models. paper
- [Arxiv, 2023] A survey of hallucination in large foundation models. 2023. paper code
- [EMNLP, 2023] Selfcheckgpt: Zero-resource black-box hallucination detection for generative large language models. 2023. paper
- [EMNLP Findings, 2021] Retrieval augmentation reduces hallucination in conversation. 2021. paper
- [Blog, 2024.11] SymptomCheck Bench. blog code
- [EMNLP, 2024] A Metric for Radiology Report Generation. paper
- [Arxiv, 2024] GMAI-MMBench: A Comprehensive Multimodal Evaluation Benchmark Towards General Medical AI. paper
- [Arxiv, 2024] Large Language Models in the Clinic: A Comprehensive Benchmark. paper code
- [Nature Reviews Bioengineering, 2023] Benchmarking medical large language models. paper
- [Bioinformatics, 2023] An extensive benchmark study on biomedical text generation and mining with ChatGPT. paper
- [Arxiv, 2023] Large language models in biomedical natural language processing: benchmarks, baselines, and recommendations. paper
- [ACL, 2023] HaluEval: A Large-Scale Hallucination Evaluation Benchmark for Large Language Models. paper code
- [ACL, 2022] Truthfulqa: Measuring how models mimic human falsehoods. paper
- [Appl. Sci, 2021] What disease does this patient have? a large-scale open domain question answering dataset from medical exams. paper
- [Arxiv, 2023] Textbooks Are All You Need. paper
- [Arxiv, 2023] Model Dementia: Generated Data Makes Models Forget. paper
- [ACL Findings, 2023] Detecting Edit Failures In Large Language Models: An Improved Specificity Benchmark. paper
- [EMNLP, 2023] Editing Large Language Models: Problems, Methods, and Opportunities. paper
- [NeurIPS, 2020] Retrieval-augmented generation for knowledge-intensive nlp tasks. paper
- [JMIR Medical Education, 2023] Differentiate ChatGPT-generated and Human-written Medical Texts. paper
- [Arxiv, 2023] Languages are rewards: Hindsight finetuning using human feedback. paper code
- [Arxiv, 2022] Training a helpful and harmless assistant with reinforcement learning from human feedback. paper code
- [Arxiv, 2022] Improving alignment of dialogue agents via targeted human judgements. paper
- [ICLR, 2021] Aligning AI with shared human values. paper code
- [Arxiv, 2021.12] Webgpt: Browser-assisted question-answering with human feedback. paper
- [Arxiv, 2023.10] A Survey of Large Language Models for Healthcare: from Data, Technology, and Applications to Accountability and Ethics. paper
- [Arxiv, 2023.8] "Do Anything Now": Characterizing and Evaluating In-The-Wild Jailbreak Prompts on Large Language Models. paper code
- [NeurIPS, 2023.7] Jailbroken: How does llm safety training fail?. paper
- [EMNLP, 2023.4] Multi-step jailbreaking privacy attacks on chatgpt. paper
- [Healthcare, 2023.3] ChatGPT utility in healthcare education, research, and practice: systematic review on the promising perspectives and valid concerns. paper
- [Nature News, 2023.1] ChatGPT listed as author on research papers: many scientists disapprove. paper
- [EMNLP, 2024.11] Large Language Models Are Poor Clinical Decision-Makers: A Comprehensive Benchmark. paper code
- [Blog, 2024.11] SymptomCheck Bench. blog code
- [Nature Communications, 2024.9] MMed-Llama3: Towards building multilingual language model for medicine. [paper] [code] [huggingface]
- [Arxiv, 2023.12] Designing Guiding Principles for NLP for Healthcare: A Case Study of Maternal Health. paper
- [JCO CCI, 2023] Natural language processing to automatically extract the presence and severity of esophagitis in notes of patients undergoing radiotherapy. [paper] [code]
- [JAMA ONC, 2023] Use of Artificial Intelligence Chatbots for Cancer Treatment Information. [paper] [code]
- [BioRxiv, 2023] A comprehensive benchmark study on biomedical text generation and mining with ChatGPT. paper
- [JAMA, 2023] Creation and adoption of large language models in medicine. paper
- [Arxiv, 2023] Large Language Models in Sport Science & Medicine: Opportunities, Risks and Considerations. paper
- [JAMA, 2023] Creation and adoption of large language models in medicine. 2023. paper
- [JAMA Forum, 2023] ChatGPT and Physicians' Malpractice Risk. paper
- [Nature Medicine, 2024] BiomedGPT A generalist vision–language foundation model for diverse biomedical tasks paper
- [Arxiv, 2023] VisionFM: a Multi-Modal Multi-Task Vision Foundation Model for Generalist Ophthalmic Artificial Intelligence. paper
- [Arxiv, 2023] A Survey on Multimodal Large Language Models. paper
- [Arxiv, 2023] Mm-react: Prompting chatgpt for multimodal reasoning and action. paper
- [Int J Oral Sci, 2023] ChatGPT for shaping the future of dentistry: the potential of multi-modal large language model. paper
- [MIDL, 2023] Frozen Language Model Helps ECG Zero-Shot Learning. paper
- [Arxiv, 2023] Exploring and Characterizing Large Language Models For Embedded System Development and Debugging. paper
- [Arxiv, 2023] The Rise and Potential of Large Language Model Based Agents: A Survey. paper
- [Arxiv, 2023] MedAgents: Large Language Models as Collaborators for Zero-shot Medical Reasoning. paper code
- [Arxiv, 2023] GeneGPT: Augmenting Large Language Models with Domain Tools for Improved Access to Biomedical Information. paper code
- [MedRxiv, 2023] OpenMedCalc: Augmentation of ChatGPT with Clinician-Informed Tools Improves Performance on Medical Calculation Tasks. paper
- [NEJM AI, 2024] Almanac — Retrieval-Augmented Language Models for Clinical Medicine. paper
- [Arxiv, 2024] ClinicalAgent: Clinical Trial Multi-Agent System with Large Language Model-based Reasoning. paper
- [Arxiv, 2024] AgentClinic: a multimodal agent benchmark to evaluate AI in simulated clinical environments paper
- [Arxiv, 2024] MDAgents: An Adaptive Collaboration of LLMs for Medical Decision-Making. paper code
- [Arxiv 2024] MediQ: Question-Asking LLMs for Adaptive and Reliable Clinical Reasoning. [paper] [code].
- LLMs Practical Guide. The codebase we built upon and it is a comprehensive LLM survey.
- Large AI Survey. Large AI Models in Health Informatics: Applications, Challenges, and the Future.
- Nature Medicine. A Survey of the Large language models in medicine.
- Healthcare LLMs Survey. A Survey of Large Language Models for Healthcare.
Please consider citing 📑 our papers if our repository is helpful to your work, thanks sincerely!
@article{zhou2023survey,
title={A Survey of Large Language Models in Medicine: Progress, Application, and Challenge},
author={Hongjian Zhou, Fenglin Liu, Boyang Gu, Xinyu Zou, Jinfa Huang, Jinge Wu, Yiru Li, Sam S. Chen, Peilin Zhou, Junling Liu, Yining Hua, Chengfeng Mao, Xian Wu, Yefeng Zheng, Lei Clifton, Zheng Li, Jiebo Luo, David A. Clifton},
journal={arXiv preprint arXiv:2311.05112},
year={2023}
}For Tasks:
Click tags to check more tools for each tasksFor Jobs:
Alternative AI tools for MedLLMsPracticalGuide
Similar Open Source Tools

MedLLMsPracticalGuide
This repository serves as a practical guide for Medical Large Language Models (Medical LLMs) and provides resources, surveys, and tools for building, fine-tuning, and utilizing LLMs in the medical domain. It covers a wide range of topics including pre-training, fine-tuning, downstream biomedical tasks, clinical applications, challenges, future directions, and more. The repository aims to provide insights into the opportunities and challenges of LLMs in medicine and serve as a practical resource for constructing effective medical LLMs.
Awesome-LLM-in-Social-Science
Awesome-LLM-in-Social-Science is a repository that compiles papers evaluating Large Language Models (LLMs) from a social science perspective. It includes papers on evaluating, aligning, and simulating LLMs, as well as enhancing tools in social science research. The repository categorizes papers based on their focus on attitudes, opinions, values, personality, morality, and more. It aims to contribute to discussions on the potential and challenges of using LLMs in social science research.

Awesome_Test_Time_LLMs
This repository focuses on test-time computing, exploring various strategies such as test-time adaptation, modifying the input, editing the representation, calibrating the output, test-time reasoning, and search strategies. It covers topics like self-supervised test-time training, in-context learning, activation steering, nearest neighbor models, reward modeling, and multimodal reasoning. The repository provides resources including papers and code for researchers and practitioners interested in enhancing the reasoning capabilities of large language models.
Fueling-Ambitions-Via-Book-Discoveries
Fueling-Ambitions-Via-Book-Discoveries is an Advanced Machine Learning & AI Course designed for students, professionals, and AI researchers. The course integrates rigorous theoretical foundations with practical coding exercises, ensuring learners develop a deep understanding of AI algorithms and their applications in finance, healthcare, robotics, NLP, cybersecurity, and more. Inspired by MIT, Stanford, and Harvard’s AI programs, it combines academic research rigor with industry-standard practices used by AI engineers at companies like Google, OpenAI, Facebook AI, DeepMind, and Tesla. Learners can learn 50+ AI techniques from top Machine Learning & Deep Learning books, code from scratch with real-world datasets, projects, and case studies, and focus on ML Engineering & AI Deployment using Django & Streamlit. The course also offers industry-relevant projects to build a strong AI portfolio.
ABigSurveyOfLLMs
ABigSurveyOfLLMs is a repository that compiles surveys on Large Language Models (LLMs) to provide a comprehensive overview of the field. It includes surveys on various aspects of LLMs such as transformers, alignment, prompt learning, data management, evaluation, societal issues, safety, misinformation, attributes of LLMs, efficient LLMs, learning methods for LLMs, multimodal LLMs, knowledge-based LLMs, extension of LLMs, LLMs applications, and more. The repository aims to help individuals quickly understand the advancements and challenges in the field of LLMs through a collection of recent surveys and research papers.
LLM-Agent-Survey
LLM-Agent-Survey is a comprehensive repository that provides a curated list of papers related to Large Language Model (LLM) agents. The repository categorizes papers based on LLM-Profiled Roles and includes high-quality publications from prestigious conferences and journals. It aims to offer a systematic understanding of LLM-based agents, covering topics such as tool use, planning, and feedback learning. The repository also includes unpublished papers with insightful analysis and novelty, marked for future updates. Users can explore a wide range of surveys, tool use cases, planning workflows, and benchmarks related to LLM agents.
ai-agent-papers
The AI Agents Papers repository provides a curated collection of papers focusing on AI agents, covering topics such as agent capabilities, applications, architectures, and presentations. It includes a variety of papers on ideation, decision making, long-horizon tasks, learning, memory-based agents, self-evolving agents, and more. The repository serves as a valuable resource for researchers and practitioners interested in AI agent technologies and advancements.
unilm
The 'unilm' repository is a collection of tools, models, and architectures for Foundation Models and General AI, focusing on tasks such as NLP, MT, Speech, Document AI, and Multimodal AI. It includes various pre-trained models, such as UniLM, InfoXLM, DeltaLM, MiniLM, AdaLM, BEiT, LayoutLM, WavLM, VALL-E, and more, designed for tasks like language understanding, generation, translation, vision, speech, and multimodal processing. The repository also features toolkits like s2s-ft for sequence-to-sequence fine-tuning and Aggressive Decoding for efficient sequence-to-sequence decoding. Additionally, it offers applications like TrOCR for OCR, LayoutReader for reading order detection, and XLM-T for multilingual NMT.
rllm
rLLM (relationLLM) is a Pytorch library for Relational Table Learning (RTL) with LLMs. It breaks down state-of-the-art GNNs, LLMs, and TNNs as standardized modules and facilitates novel model building in a 'combine, align, and co-train' way using these modules. The library is LLM-friendly, processes various graphs as multiple tables linked by foreign keys, introduces new relational table datasets, and is supported by students and teachers from Shanghai Jiao Tong University and Tsinghua University.
LLM-on-Tabular-Data-Prediction-Table-Understanding-Data-Generation
This repository serves as a comprehensive survey on the application of Large Language Models (LLMs) on tabular data, focusing on tasks such as prediction, data generation, and table understanding. It aims to consolidate recent progress in this field by summarizing key techniques, metrics, datasets, models, and optimization approaches. The survey identifies strengths, limitations, unexplored territories, and gaps in the existing literature, providing insights for future research directions. It also offers code and dataset references to empower readers with the necessary tools and knowledge to address challenges in this rapidly evolving domain.
OpenRedTeaming
OpenRedTeaming is a repository focused on red teaming for generative models, specifically large language models (LLMs). The repository provides a comprehensive survey on potential attacks on GenAI and robust safeguards. It covers attack strategies, evaluation metrics, benchmarks, and defensive approaches. The repository also implements over 30 auto red teaming methods. It includes surveys, taxonomies, attack strategies, and risks related to LLMs. The goal is to understand vulnerabilities and develop defenses against adversarial attacks on large language models.
LLM-Tool-Survey
This repository contains a collection of papers related to tool learning with large language models (LLMs). The papers are organized according to the survey paper 'Tool Learning with Large Language Models: A Survey'. The survey focuses on the benefits and implementation of tool learning with LLMs, covering aspects such as task planning, tool selection, tool calling, response generation, benchmarks, evaluation, challenges, and future directions in the field. It aims to provide a comprehensive understanding of tool learning with LLMs and inspire further exploration in this emerging area.
Awesome-LLM-Interpretability
Awesome-LLM-Interpretability is a curated list of materials related to LLM (Large Language Models) interpretability, covering tutorials, code libraries, surveys, videos, papers, and blogs. It includes resources on transformer mechanistic interpretability, visualization, interventions, probing, fine-tuning, feature representation, learning dynamics, knowledge editing, hallucination detection, and redundancy analysis. The repository aims to provide a comprehensive overview of tools, techniques, and methods for understanding and interpreting the inner workings of large language models.
awesome-azure-openai-llm
This repository is a collection of references to Azure OpenAI, Large Language Models (LLM), and related services and libraries. It provides information on various topics such as RAG, Azure OpenAI, LLM applications, agent design patterns, semantic kernel, prompting, finetuning, challenges & abilities, LLM landscape, surveys & references, AI tools & extensions, datasets, and evaluations. The content covers a wide range of topics related to AI, machine learning, and natural language processing, offering insights into the latest advancements in the field.
Awesome-Embodied-Agent-with-LLMs
This repository, named Awesome-Embodied-Agent-with-LLMs, is a curated list of research related to Embodied AI or agents with Large Language Models. It includes various papers, surveys, and projects focusing on topics such as self-evolving agents, advanced agent applications, LLMs with RL or world models, planning and manipulation, multi-agent learning and coordination, vision and language navigation, detection, 3D grounding, interactive embodied learning, rearrangement, benchmarks, simulators, and more. The repository provides a comprehensive collection of resources for individuals interested in exploring the intersection of embodied agents and large language models.
Awesome_papers_on_LLMs_detection
This repository is a curated list of papers focused on the detection of Large Language Models (LLMs)-generated content. It includes the latest research papers covering detection methods, datasets, attacks, and more. The repository is regularly updated to include the most recent papers in the field.
For similar tasks
Detection-and-Classification-of-Alzheimers-Disease
This tool is designed to detect and classify Alzheimer's Disease using Deep Learning and Machine Learning algorithms on an early basis, which is further optimized using the Crow Search Algorithm (CSA). Alzheimer's is a fatal disease, and early detection is crucial for patients to predetermine their condition and prevent its progression. By analyzing MRI scanned images using Artificial Intelligence technology, this tool can classify patients who may or may not develop AD in the future. The CSA algorithm, combined with ML algorithms, has proven to be the most effective approach for this purpose.

MedLLMsPracticalGuide
This repository serves as a practical guide for Medical Large Language Models (Medical LLMs) and provides resources, surveys, and tools for building, fine-tuning, and utilizing LLMs in the medical domain. It covers a wide range of topics including pre-training, fine-tuning, downstream biomedical tasks, clinical applications, challenges, future directions, and more. The repository aims to provide insights into the opportunities and challenges of LLMs in medicine and serve as a practical resource for constructing effective medical LLMs.

AMIE-pytorch
Implementation of the general framework for AMIE, from the paper Towards Conversational Diagnostic AI, out of Google Deepmind. This repository provides a Pytorch implementation of the AMIE framework, aimed at enabling conversational diagnostic AI. It is a work in progress and welcomes collaboration from individuals with a background in deep learning and an interest in medical applications.
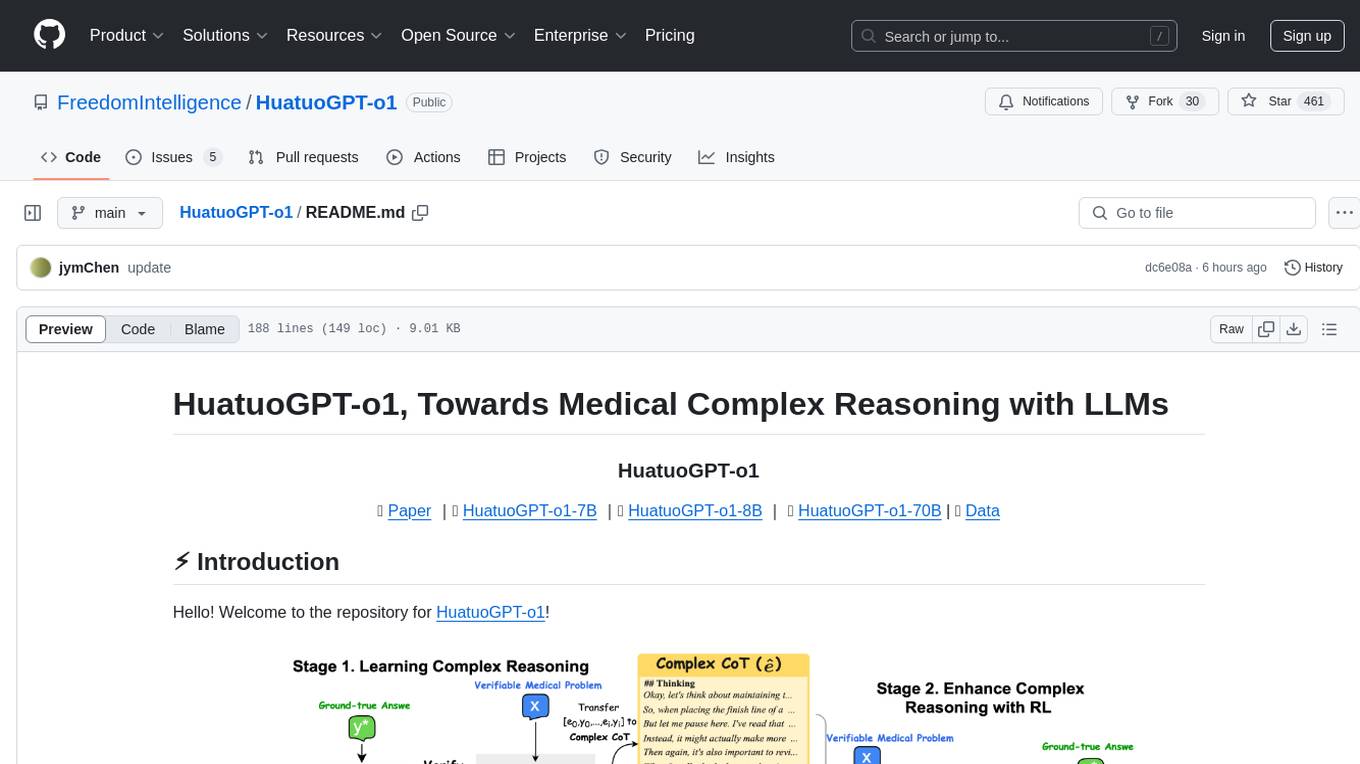
HuatuoGPT-o1
HuatuoGPT-o1 is a medical language model designed for advanced medical reasoning. It can identify mistakes, explore alternative strategies, and refine answers. The model leverages verifiable medical problems and a specialized medical verifier to guide complex reasoning trajectories and enhance reasoning through reinforcement learning. The repository provides access to models, data, and code for HuatuoGPT-o1, allowing users to deploy the model for medical reasoning tasks.
minions
Minions is a communication protocol that enables small on-device models to collaborate with frontier models in the cloud. By only reading long contexts locally, it reduces cloud costs with minimal or no quality degradation. The repository provides a demonstration of the protocol.
For similar jobs

MedLLMsPracticalGuide
This repository serves as a practical guide for Medical Large Language Models (Medical LLMs) and provides resources, surveys, and tools for building, fine-tuning, and utilizing LLMs in the medical domain. It covers a wide range of topics including pre-training, fine-tuning, downstream biomedical tasks, clinical applications, challenges, future directions, and more. The repository aims to provide insights into the opportunities and challenges of LLMs in medicine and serve as a practical resource for constructing effective medical LLMs.
Taiyi-LLM
Taiyi (太一) is a bilingual large language model fine-tuned for diverse biomedical tasks. It aims to facilitate communication between healthcare professionals and patients, provide medical information, and assist in diagnosis, biomedical knowledge discovery, drug development, and personalized healthcare solutions. The model is based on the Qwen-7B-base model and has been fine-tuned using rich bilingual instruction data. It covers tasks such as question answering, biomedical dialogue, medical report generation, biomedical information extraction, machine translation, title generation, text classification, and text semantic similarity. The project also provides standardized data formats, model training details, model inference guidelines, and overall performance metrics across various BioNLP tasks.
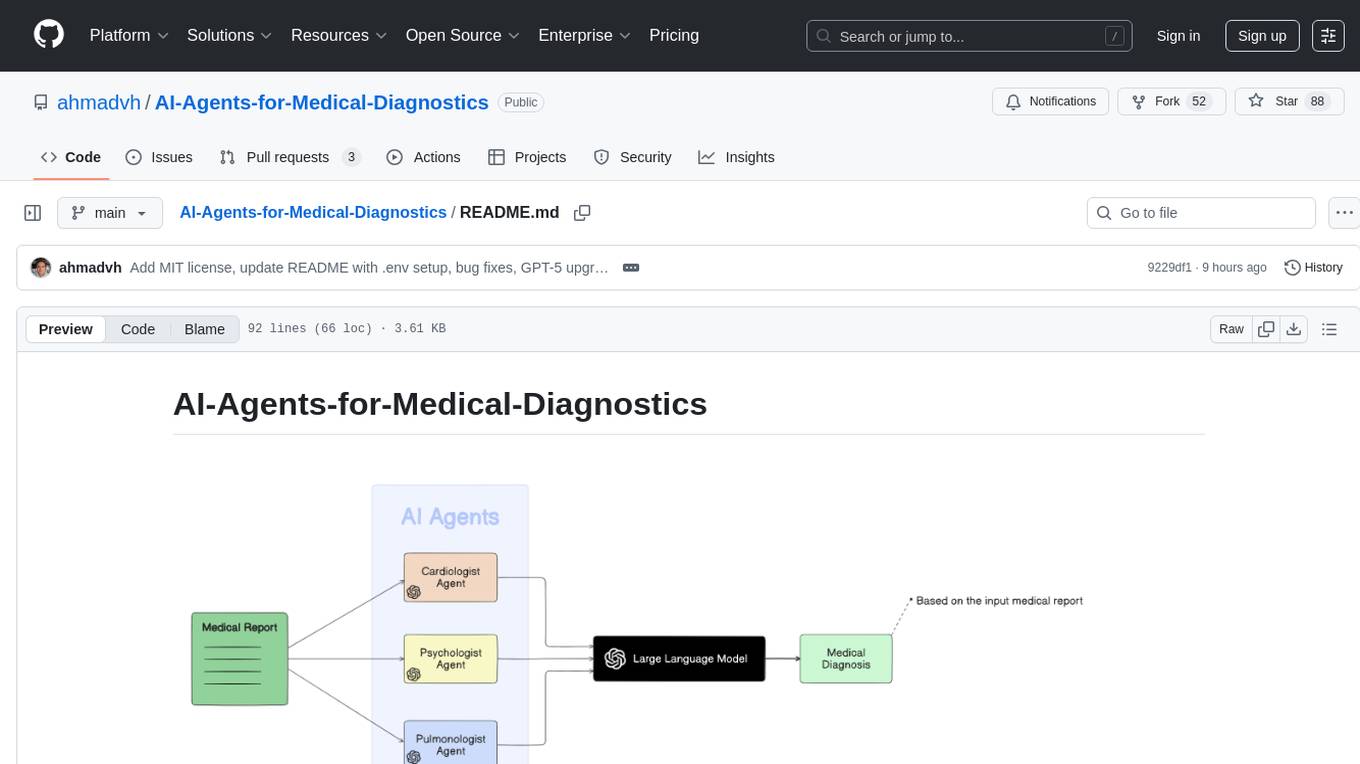
AI-Agents-for-Medical-Diagnostics
AI Agents for Medical Diagnostics is a repository containing a collection of machine learning models and algorithms designed to assist in medical diagnosis. The tools provided in this repository are specifically tailored for analyzing medical data and making predictions related to various health conditions. By leveraging the power of artificial intelligence, these agents aim to improve the accuracy and efficiency of diagnostic processes in the medical field. Researchers, healthcare professionals, and data scientists can benefit from the resources available in this repository to develop innovative solutions for diagnosing illnesses and predicting patient outcomes.
fuse-med-ml
FuseMedML is a Python framework designed to accelerate machine learning-based discovery in the medical field by promoting code reuse. It provides a flexible design concept where data is stored in a nested dictionary, allowing easy handling of multi-modality information. The framework includes components for creating custom models, loss functions, metrics, and data processing operators. Additionally, FuseMedML offers 'batteries included' key components such as fuse.data for data processing, fuse.eval for model evaluation, and fuse.dl for reusable deep learning components. It supports PyTorch and PyTorch Lightning libraries and encourages the creation of domain extensions for specific medical domains.
hi-ml
The Microsoft Health Intelligence Machine Learning Toolbox is a repository that provides low-level and high-level building blocks for Machine Learning / AI researchers and practitioners. It simplifies and streamlines work on deep learning models for healthcare and life sciences by offering tested components such as data loaders, pre-processing tools, deep learning models, and cloud integration utilities. The repository includes two Python packages, 'hi-ml-azure' for helper functions in AzureML, 'hi-ml' for ML components, and 'hi-ml-cpath' for models and workflows related to histopathology images.
SlicerTotalSegmentator
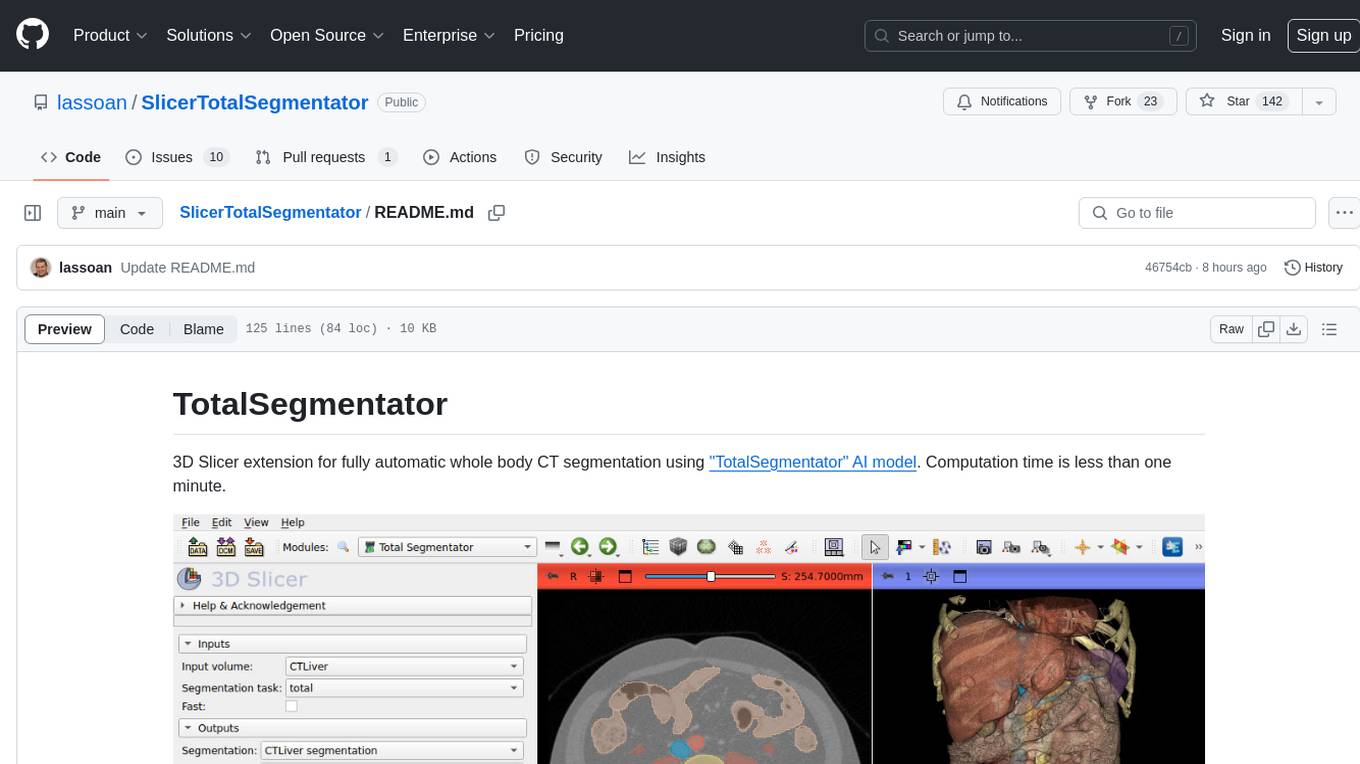
TotalSegmentator is a 3D Slicer extension designed for fully automatic whole body CT segmentation using the 'TotalSegmentator' AI model. The computation time is less than one minute, making it efficient for research purposes. Users can set up GPU acceleration for faster segmentation. The tool provides a user-friendly interface for loading CT images, creating segmentations, and displaying results in 3D. Troubleshooting steps are available for common issues such as failed computation, GPU errors, and inaccurate segmentations. Contributions to the extension are welcome, following 3D Slicer contribution guidelines.
machine-learning-research
The 'machine-learning-research' repository is a comprehensive collection of resources related to mathematics, machine learning, deep learning, artificial intelligence, data science, and various scientific fields. It includes materials such as courses, tutorials, books, podcasts, communities, online courses, papers, and dissertations. The repository covers topics ranging from fundamental math skills to advanced machine learning concepts, with a focus on applications in healthcare, genetics, computational biology, precision health, and AI in science. It serves as a valuable resource for individuals interested in learning and researching in the fields of machine learning and related disciplines.
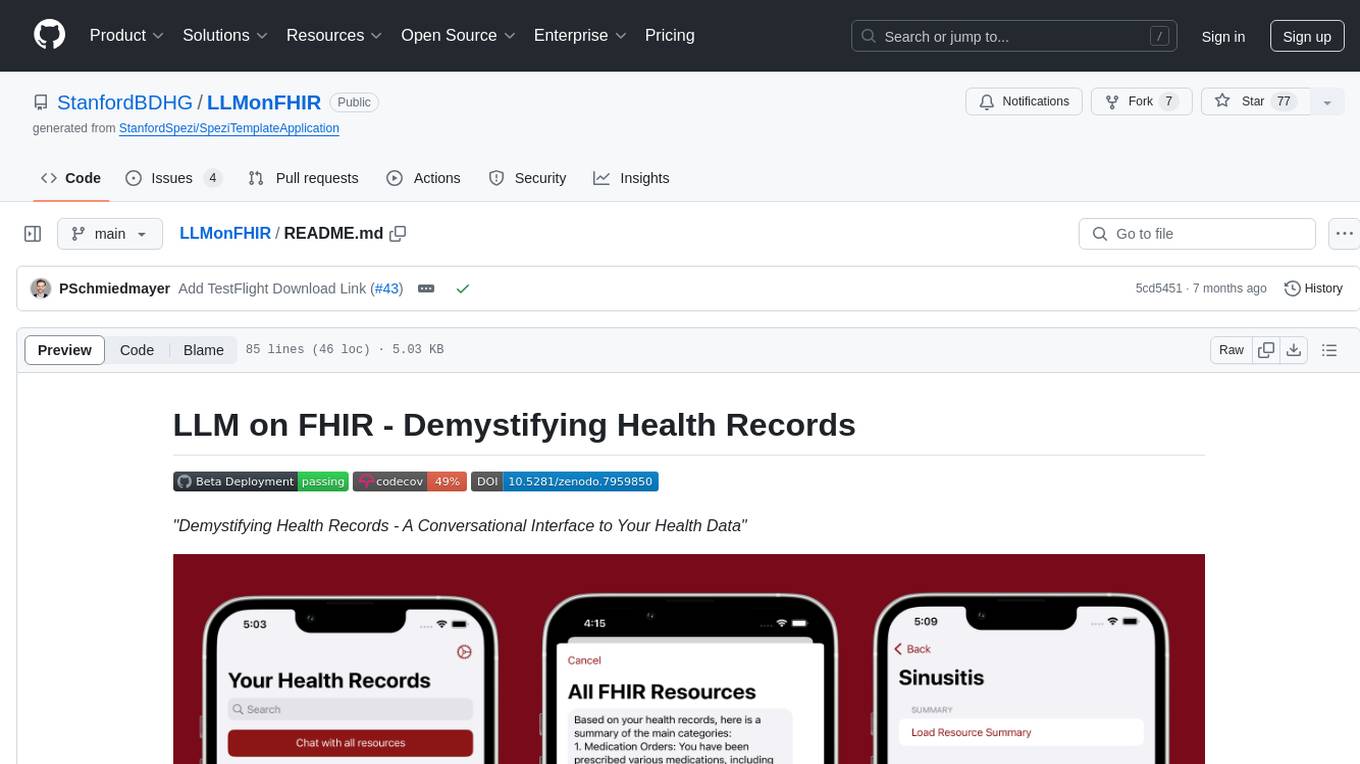
LLMonFHIR
LLMonFHIR is an iOS application that utilizes large language models (LLMs) to interpret and provide context around patient data in the Fast Healthcare Interoperability Resources (FHIR) format. It connects to the OpenAI GPT API to analyze FHIR resources, supports multiple languages, and allows users to interact with their health data stored in the Apple Health app. The app aims to simplify complex health records, provide insights, and facilitate deeper understanding through a conversational interface. However, it is an experimental app for informational purposes only and should not be used as a substitute for professional medical advice. Users are advised to verify information provided by AI models and consult healthcare professionals for personalized advice.













