
ProLLM
[COLM'24] We propose Protein Chain of Thought (ProCoT), which replicates the biological mechanism of signaling pathways as language prompts. It considers a signaling pathway as a protein reasoning process, which starts from upstream proteins and passes through several intermediate proteins to transmit biological signals to downstream proteins.
Stars: 58
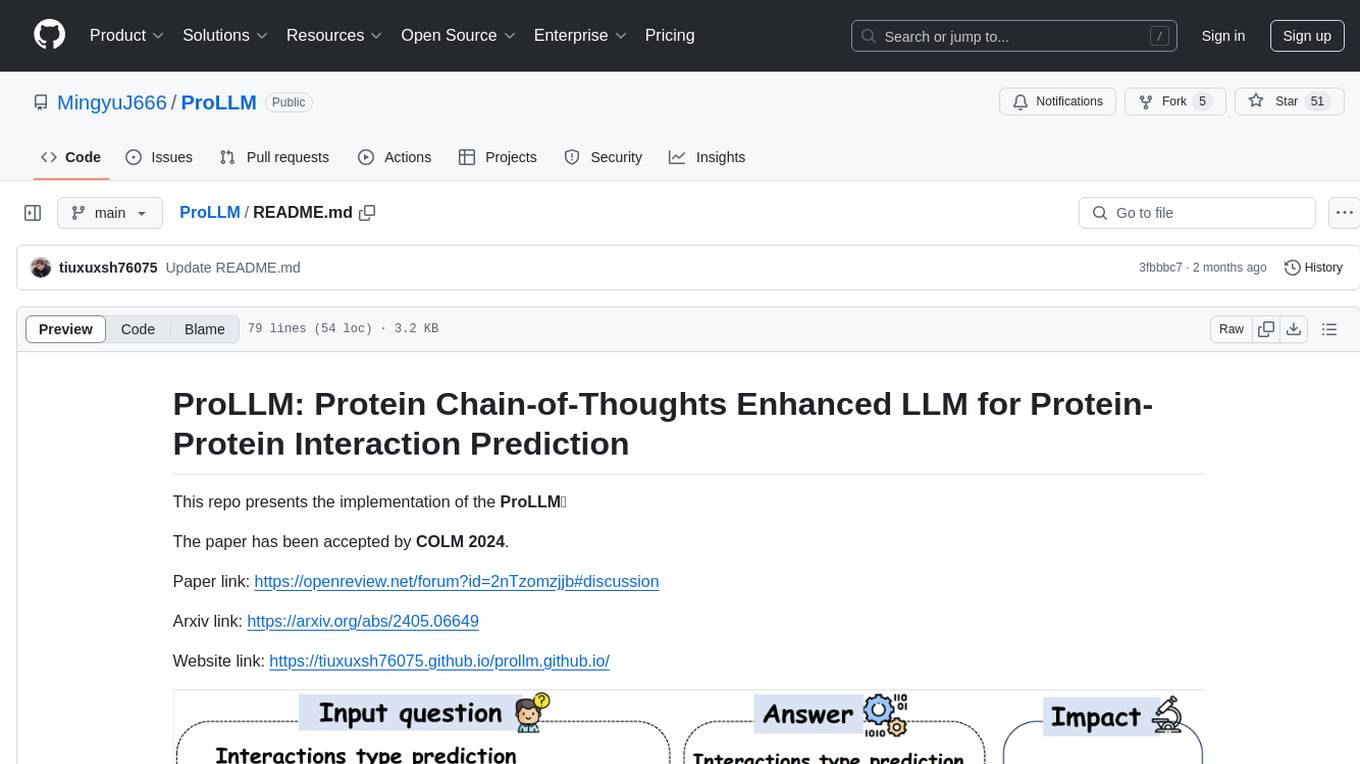
ProLLM is a framework that leverages Large Language Models to interpret and analyze protein sequences and interactions through natural language processing. It introduces the Protein Chain of Thought (ProCoT) method to transform complex protein interaction data into intuitive prompts, enhancing predictive accuracy by incorporating protein-specific embeddings and fine-tuning on domain-specific datasets.
README:
This repo presents the implementation of the ProLLM🧬
The paper has been accepted by COLM 2024.
Paper link: https://openreview.net/forum?id=2nTzomzjjb#discussion
Arxiv link: https://arxiv.org/abs/2405.06649
Website link: https://tiuxuxsh76075.github.io/prollm.github.io/
We present Protein Chain-of-Thoughts Enhanced LLM for Protein-Protein Interaction Prediction, abbreviated as ProLLM. This innovative framework leverages the advanced capabilities of Large Language Models (LLMs) to interpret and analyze protein sequences and interactions through a natural language processing lens.
-
Protein Chain of Thought (ProCoT) Method: ProLLM introduces the Protein Chain of Thought (ProCoT) method, transforming the complex, structured data of protein interactions into intuitive, natural language prompts.
-
Enhanced Predictive Accuracy: This approach not only facilitates a deeper understanding of protein functions and interactions but also enhances the model's predictive accuracy by incorporating protein-specific embeddings and instruction fine-tuning on domain-specific datasets.
pip install -r requirements.txt
- Download SHS27K, SHS148K, STRING and Human
- Preprocess the dataset into Protein Chain of Thought (ProCoT) by the ./data_preprocess/[dataset_name]/preprocess.py
python [dataset]_preprocess.py
- Do the embedding replacement in ./embedding_replacement (Embedding needs to be run on your own, as it is too large to be uploaded.)
- Instruction fintuning in ./Instruction_fintuning
- Do training through train_ProLLM.py , make sure the location of model and tokenizer in right place. Feel free to change 'num_epochs', 'batch_size' and 'learning_rate'.
python train_ProLLM.py --model_dir /path/to/model/dir --tokenizer_dir /path/to/tokenizer/dir --data_file dataset.csv --num_epochs 1 --batch_size 2 --learning_rate 3e-4 --output_dir /path/to/output/dir
If you find this paper helpful, please consider to cite:
@inproceedings{
jin2024prollm,
title={Pro{LLM}: Protein Chain-of-Thoughts Enhanced {LLM} for Protein-Protein Interaction Prediction},
author={Mingyu Jin and Haochen Xue and Zhenting Wang and Boming Kang and Ruosong Ye and Kaixiong Zhou and Mengnan Du and Yongfeng Zhang},
booktitle={First Conference on Language Modeling},
year={2024},
url={https://openreview.net/forum?id=2nTzomzjjb}
}
@article{jin2024prollm,
title={ProLLM: Protein Chain-of-Thoughts Enhanced LLM for Protein-Protein Interaction Prediction},
author={Jin, Mingyu and Xue, Haochen and Wang, Zhenting and Kang, Boming and Ye, Ruosong and Zhou, Kaixiong and Du, Mengnan and Zhang, Yongfeng},
journal={arXiv e-prints},
pages={arXiv--2405},
year={2024}
}
For Tasks:
Click tags to check more tools for each tasksFor Jobs:
Alternative AI tools for ProLLM
Similar Open Source Tools

ProLLM
ProLLM is a framework that leverages Large Language Models to interpret and analyze protein sequences and interactions through natural language processing. It introduces the Protein Chain of Thought (ProCoT) method to transform complex protein interaction data into intuitive prompts, enhancing predictive accuracy by incorporating protein-specific embeddings and fine-tuning on domain-specific datasets.

only_train_once
Only Train Once (OTO) is an automatic, architecture-agnostic DNN training and compression framework that allows users to train a general DNN from scratch or a pretrained checkpoint to achieve high performance and slimmer architecture simultaneously in a one-shot manner without fine-tuning. The framework includes features for automatic structured pruning and erasing operators, as well as hybrid structured sparse optimizers for efficient model compression. OTO provides tools for pruning zero-invariant group partitioning, constructing pruned models, and visualizing pruning and erasing dependency graphs. It supports the HESSO optimizer and offers a sanity check for compliance testing on various DNNs. The repository also includes publications, installation instructions, quick start guides, and a roadmap for future enhancements and collaborations.
Macaw-LLM
Macaw-LLM is a pioneering multi-modal language modeling tool that seamlessly integrates image, audio, video, and text data. It builds upon CLIP, Whisper, and LLaMA models to process and analyze multi-modal information effectively. The tool boasts features like simple and fast alignment, one-stage instruction fine-tuning, and a new multi-modal instruction dataset. It enables users to align multi-modal features efficiently, encode instructions, and generate responses across different data types.
Genesis
Genesis is a physics platform designed for general purpose Robotics/Embodied AI/Physical AI applications. It includes a universal physics engine, a lightweight, ultra-fast, pythonic, and user-friendly robotics simulation platform, a powerful and fast photo-realistic rendering system, and a generative data engine that transforms user-prompted natural language description into various modalities of data. It aims to lower the barrier to using physics simulations, unify state-of-the-art physics solvers, and minimize human effort in collecting and generating data for robotics and other domains.
Reflection_Tuning
Reflection-Tuning is a project focused on improving the quality of instruction-tuning data through a reflection-based method. It introduces Selective Reflection-Tuning, where the student model can decide whether to accept the improvements made by the teacher model. The project aims to generate high-quality instruction-response pairs by defining specific criteria for the oracle model to follow and respond to. It also evaluates the efficacy and relevance of instruction-response pairs using the r-IFD metric. The project provides code for reflection and selection processes, along with data and model weights for both V1 and V2 methods.
nous
Nous is an open-source TypeScript platform for autonomous AI agents and LLM based workflows. It aims to automate processes, support requests, review code, assist with refactorings, and more. The platform supports various integrations, multiple LLMs/services, CLI and web interface, human-in-the-loop interactions, flexible deployment options, observability with OpenTelemetry tracing, and specific agents for code editing, software engineering, and code review. It offers advanced features like reasoning/planning, memory and function call history, hierarchical task decomposition, and control-loop function calling options. Nous is designed to be a flexible platform for the TypeScript community to expand and support different use cases and integrations.
FATE-LLM
FATE-LLM is a framework supporting federated learning for large and small language models. It promotes training efficiency of federated LLMs using Parameter-Efficient methods, protects the IP of LLMs using FedIPR, and ensures data privacy during training and inference through privacy-preserving mechanisms.

raga-llm-hub
Raga LLM Hub is a comprehensive evaluation toolkit for Language and Learning Models (LLMs) with over 100 meticulously designed metrics. It allows developers and organizations to evaluate and compare LLMs effectively, establishing guardrails for LLMs and Retrieval Augmented Generation (RAG) applications. The platform assesses aspects like Relevance & Understanding, Content Quality, Hallucination, Safety & Bias, Context Relevance, Guardrails, and Vulnerability scanning, along with Metric-Based Tests for quantitative analysis. It helps teams identify and fix issues throughout the LLM lifecycle, revolutionizing reliability and trustworthiness.
ChatLaw
ChatLaw is an open-source legal large language model tailored for Chinese legal scenarios. It aims to combine LLM and knowledge bases to provide solutions for legal scenarios. The models include ChatLaw-13B and ChatLaw-33B, trained on various legal texts to construct dialogue data. The project focuses on improving logical reasoning abilities and plans to train models with parameters exceeding 30B for better performance. The dataset consists of forum posts, news, legal texts, judicial interpretations, legal consultations, exam questions, and court judgments, cleaned and enhanced to create dialogue data. The tool is designed to assist in legal tasks requiring complex logical reasoning, with a focus on accuracy and reliability.
FinRobot
FinRobot is an open-source AI agent platform designed for financial applications using large language models. It transcends the scope of FinGPT, offering a comprehensive solution that integrates a diverse array of AI technologies. The platform's versatility and adaptability cater to the multifaceted needs of the financial industry. FinRobot's ecosystem is organized into four layers, including Financial AI Agents Layer, Financial LLMs Algorithms Layer, LLMOps and DataOps Layers, and Multi-source LLM Foundation Models Layer. The platform's agent workflow involves Perception, Brain, and Action modules to capture, process, and execute financial data and insights. The Smart Scheduler optimizes model diversity and selection for tasks, managed by components like Director Agent, Agent Registration, Agent Adaptor, and Task Manager. The tool provides a structured file organization with subfolders for agents, data sources, and functional modules, along with installation instructions and hands-on tutorials.
InstructGraph
InstructGraph is a framework designed to enhance large language models (LLMs) for graph-centric tasks by utilizing graph instruction tuning and preference alignment. The tool collects and decomposes 29 standard graph datasets into four groups, enabling LLMs to better understand and generate graph data. It introduces a structured format verbalizer to transform graph data into a code-like format, facilitating code understanding and generation. Additionally, it addresses hallucination problems in graph reasoning and generation through direct preference optimization (DPO). The tool aims to bridge the gap between textual LLMs and graph data, offering a comprehensive solution for graph-related tasks.
Vision-LLM-Alignment
Vision-LLM-Alignment is a repository focused on implementing alignment training for visual large language models (LLMs), including SFT training, reward model training, and PPO/DPO training. It supports various model architectures and provides datasets for training. The repository also offers benchmark results and installation instructions for users.
pytorch-forecasting
PyTorch Forecasting is a PyTorch-based package for time series forecasting with state-of-the-art network architectures. It offers a high-level API for training networks on pandas data frames and utilizes PyTorch Lightning for scalable training on GPUs and CPUs. The package aims to simplify time series forecasting with neural networks by providing a flexible API for professionals and default settings for beginners. It includes a timeseries dataset class, base model class, multiple neural network architectures, multi-horizon timeseries metrics, and hyperparameter tuning with optuna. PyTorch Forecasting is built on pytorch-lightning for easy training on various hardware configurations.
MMC
This repository, MMC, focuses on advancing multimodal chart understanding through large-scale instruction tuning. It introduces a dataset supporting various tasks and chart types, a benchmark for evaluating reasoning capabilities over charts, and an assistant achieving state-of-the-art performance on chart QA benchmarks. The repository provides data for chart-text alignment, benchmarking, and instruction tuning, along with existing datasets used in experiments. Additionally, it offers a Gradio demo for the MMCA model.
gemma
Gemma is a family of open-weights Large Language Model (LLM) by Google DeepMind, based on Gemini research and technology. This repository contains an inference implementation and examples, based on the Flax and JAX frameworks. Gemma can run on CPU, GPU, and TPU, with model checkpoints available for download. It provides tutorials, reference implementations, and Colab notebooks for tasks like sampling and fine-tuning. Users can contribute to Gemma through bug reports and pull requests. The code is licensed under the Apache License, Version 2.0.
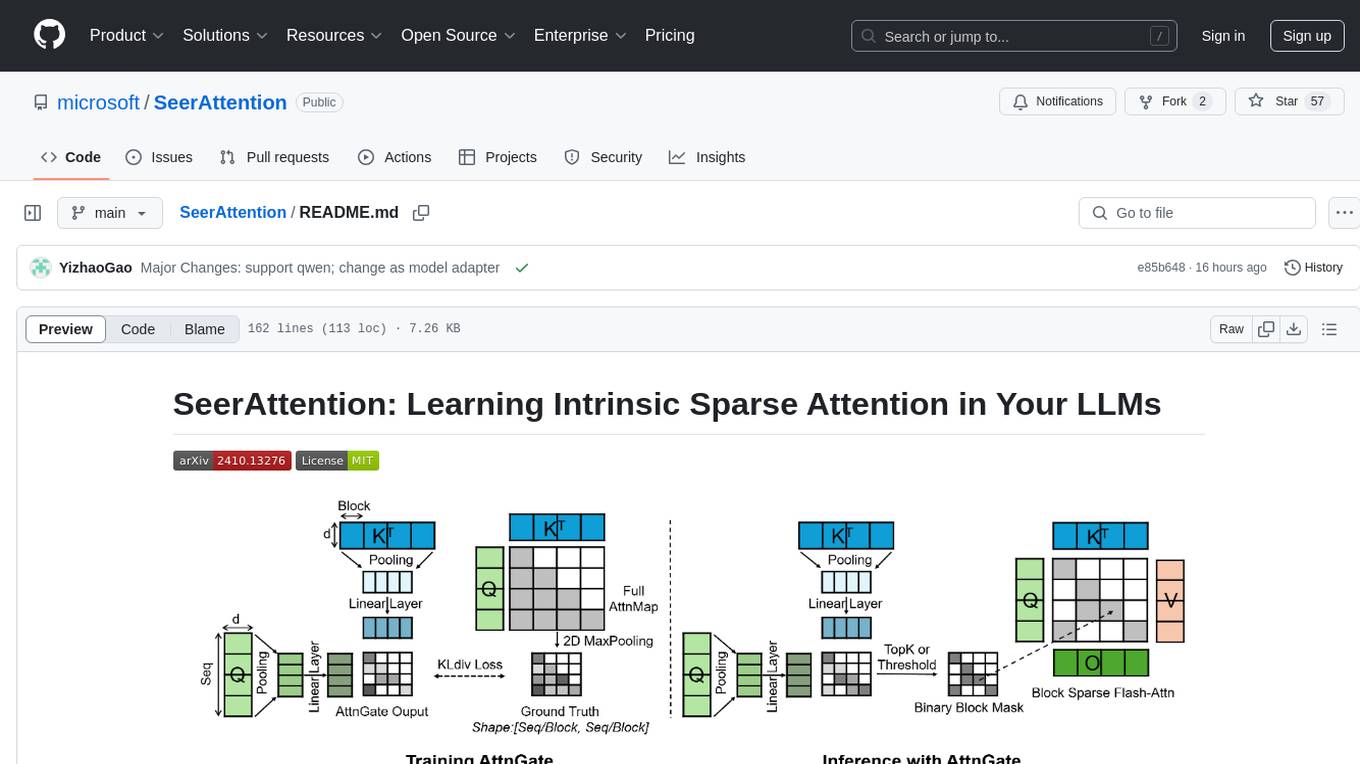
SeerAttention
SeerAttention is a novel trainable sparse attention mechanism that learns intrinsic sparsity patterns directly from LLMs through self-distillation at post-training time. It achieves faster inference while maintaining accuracy for long-context prefilling. The tool offers features such as trainable sparse attention, block-level sparsity, self-distillation, efficient kernel, and easy integration with existing transformer architectures. Users can quickly start using SeerAttention for inference with AttnGate Adapter and training attention gates with self-distillation. The tool provides efficient evaluation methods and encourages contributions from the community.
For similar tasks

ProLLM
ProLLM is a framework that leverages Large Language Models to interpret and analyze protein sequences and interactions through natural language processing. It introduces the Protein Chain of Thought (ProCoT) method to transform complex protein interaction data into intuitive prompts, enhancing predictive accuracy by incorporating protein-specific embeddings and fine-tuning on domain-specific datasets.
TokenPacker
TokenPacker is a novel visual projector that compresses visual tokens by 75%∼89% with high efficiency. It adopts a 'coarse-to-fine' scheme to generate condensed visual tokens, achieving comparable or better performance across diverse benchmarks. The tool includes TokenPacker for general use and TokenPacker-HD for high-resolution image understanding. It provides training scripts, checkpoints, and supports various compression ratios and patch numbers.

Macaw-LLM
Macaw-LLM is a pioneering multi-modal language modeling tool that seamlessly integrates image, audio, video, and text data. It builds upon CLIP, Whisper, and LLaMA models to process and analyze multi-modal information effectively. The tool boasts features like simple and fast alignment, one-stage instruction fine-tuning, and a new multi-modal instruction dataset. It enables users to align multi-modal features efficiently, encode instructions, and generate responses across different data types.
For similar jobs
Generative-AI-Drug-Discovery
Generative-AI-Drug-Discovery is a public repository on GitHub focused on using tensor network machine learning approaches to accelerate GenAI for drug discovery. The repository aims to implement effective architectures and methodologies into Large Language Models (LLMs) to enhance Drug Discovery Generative AI performance.
AI2BMD
AI2BMD is a program for efficiently simulating protein molecular dynamics with ab initio accuracy. The repository contains datasets, simulation programs, and public materials related to AI2BMD. It provides a Docker image for easy deployment and a standalone launcher program. Users can run simulations by downloading the launcher script and specifying simulation parameters. The repository also includes ready-to-use protein structures for testing. AI2BMD is designed for x86-64 GNU/Linux systems with recommended hardware specifications. The related research includes model architectures like ViSNet, Geoformer, and fine-grained force metrics for MLFF. Citation information and contact details for the AI2BMD Team are provided.
mercure
mercure DICOM Orchestrator is a flexible solution for routing and processing DICOM files. It offers a user-friendly web interface and extensive monitoring functions. Custom processing modules can be implemented as Docker containers. Written in Python, it uses the DCMTK toolkit for DICOM communication. It can be deployed as a single-server installation using Docker Compose or as a scalable cluster installation using Nomad. mercure consists of service modules for receiving, routing, processing, dispatching, cleaning, web interface, and central monitoring.

ProLLM
ProLLM is a framework that leverages Large Language Models to interpret and analyze protein sequences and interactions through natural language processing. It introduces the Protein Chain of Thought (ProCoT) method to transform complex protein interaction data into intuitive prompts, enhancing predictive accuracy by incorporating protein-specific embeddings and fine-tuning on domain-specific datasets.
grand-challenge.org
Grand Challenge is a platform that provides access to large amounts of annotated training data, objective comparisons of state-of-the-art machine learning solutions, and clinical validation using real-world data. It assists researchers, data scientists, and clinicians in collaborating to develop robust machine learning solutions to problems in biomedical imaging.
Detection-and-Classification-of-Alzheimers-Disease
This tool is designed to detect and classify Alzheimer's Disease using Deep Learning and Machine Learning algorithms on an early basis, which is further optimized using the Crow Search Algorithm (CSA). Alzheimer's is a fatal disease, and early detection is crucial for patients to predetermine their condition and prevent its progression. By analyzing MRI scanned images using Artificial Intelligence technology, this tool can classify patients who may or may not develop AD in the future. The CSA algorithm, combined with ML algorithms, has proven to be the most effective approach for this purpose.

OpenCRISPR
OpenCRISPR is a set of free and open gene editing systems designed by Profluent Bio. The OpenCRISPR-1 protein maintains the prototypical architecture of a Type II Cas9 nuclease but is hundreds of mutations away from SpCas9 or any other known natural CRISPR-associated protein. You can view OpenCRISPR-1 as a drop-in replacement for many protocols that need a cas9-like protein with an NGG PAM and you can even use it with canonical SpCas9 gRNAs. OpenCRISPR-1 can be fused in a deactivated or nickase format for next generation gene editing techniques like base, prime, or epigenome editing.

AlphaFold3
AlphaFold3 is an implementation of the Alpha Fold 3 model in PyTorch for accurate structure prediction of biomolecular interactions. It includes modules for genetic diffusion and full model examples for forward pass computations. The tool allows users to generate random pair and single representations, operate on atomic coordinates, and perform structure predictions based on input tensors. The implementation also provides functionalities for training and evaluating the model.

