
aioli
Framework for building fast genomics web tools with WebAssembly and WebWorkers
Stars: 100
Aioli is a library for running genomics command-line tools in the browser using WebAssembly. It creates a single WebWorker to run all WebAssembly tools, shares a filesystem across modules, and efficiently mounts local files. The tool encapsulates each module for loading, does WebAssembly feature detection, and communicates with the WebWorker using the Comlink library. Users can deploy new releases and versions, and benefit from code reuse by porting existing C/C++/Rust/etc tools to WebAssembly for browser use.
README:
Aioli is a library for running genomics command-line tools in the browser using WebAssembly. See Who uses biowasm for example use cases.
Run npm install to install dependencies.
Then run npm run dev to launch the web server and use src/example.js as a sample web app that uses the dev version of Aioli.
Run npm run test.
- Update version in
package.json(append-rc1for release candidates) - Build:
npm run build - Create npm package:
npm pack - Publish package:
npm publish [tgzfile] --tag next - To use pre-release version:
npm install @biowasm/aioli@next
- Update version in
package.json - Build:
npm run build - Publish package:
npm publish --access public - Create release in the GitHub repo
- Add to
biowasm.json - Deploy biowasm CDN with env=stg, tool=aioli and run biowasm tests to validate that stg tests still pass
- Repeat with env=prd
- Aioli creates a single WebWorker, in which all WebAssembly tools run.
- We use a PROXYFS virtual filesystem so we can share a filesystem across all WebAssembly modules, i.e. the output of one tool can be used as the input of another tool.
- We use a WORKERFS virtual filesystem to mount local files efficiently (i.e. without having to load all their contents into memory). To support use cases where tools need to create files in the same folder as those ready-only files (e.g.
samtools index), we automatically create a symlink from each local file's WORKERFS path to a path in PROXYFS. - Once the WebWorker initializes, it loads the WebAssembly modules one at a time. To support this, we need to encapsulate each module using Emscripten's
-s MODULARIZE=1, i.e. the.jsfile will contain aModulefunction that initializes the module and returns aPromisethat resolves when the module is loaded. - We do WebAssembly feature detection at initialization using
wasm-feature-detect. If a tool needs WebAssembly SIMD and the user has a browser that does not support it, we will load the non-SIMD version of that tool. - We communicate with the WebWorker using the Comlink library.
WebAssembly is a fast, low-level, compiled binary instruction format that runs in all major browsers at near native speeds. One key feature of WebAssembly is code reuse: you can port existing C/C++/Rust/etc tools to WebAssembly so those tools can run in the browser.
WebWorkers allow you to run JavaScript in the browser in a background thread, which keeps the browser responsive.
See the biowasm project.
For Tasks:
Click tags to check more tools for each tasksFor Jobs:
Alternative AI tools for aioli
Similar Open Source Tools

aioli
Aioli is a library for running genomics command-line tools in the browser using WebAssembly. It creates a single WebWorker to run all WebAssembly tools, shares a filesystem across modules, and efficiently mounts local files. The tool encapsulates each module for loading, does WebAssembly feature detection, and communicates with the WebWorker using the Comlink library. Users can deploy new releases and versions, and benefit from code reuse by porting existing C/C++/Rust/etc tools to WebAssembly for browser use.
desktop
ComfyUI Desktop is a packaged desktop application that allows users to easily use ComfyUI with bundled features like ComfyUI source code, ComfyUI-Manager, and uv. It automatically installs necessary Python dependencies and updates with stable releases. The app comes with Electron, Chromium binaries, and node modules. Users can store ComfyUI files in a specified location and manage model paths. The tool requires Python 3.12+ and Visual Studio with Desktop C++ workload for Windows. It uses nvm to manage node versions and yarn as the package manager. Users can install ComfyUI and dependencies using comfy-cli, download uv, and build/launch the code. Troubleshooting steps include rebuilding modules and installing missing libraries. The tool supports debugging in VSCode and provides utility scripts for cleanup. Crash reports can be sent to help debug issues, but no personal data is included.
starter-monorepo
Starter Monorepo is a template repository for setting up a monorepo structure in your project. It provides a basic setup with configurations for managing multiple packages within a single repository. This template includes tools for package management, versioning, testing, and deployment. By using this template, you can streamline your development process, improve code sharing, and simplify dependency management across your project. Whether you are working on a small project or a large-scale application, Starter Monorepo can help you organize your codebase efficiently and enhance collaboration among team members.
agnai
Agnaistic is an AI roleplay chat tool that allows users to interact with personalized characters using their favorite AI services. It supports multiple AI services, persona schema formats, and features such as group conversations, user authentication, and memory/lore books. Agnaistic can be self-hosted or run using Docker, and it provides a range of customization options through its settings.json file. The tool is designed to be user-friendly and accessible, making it suitable for both casual users and developers.

unitycatalog
Unity Catalog is an open and interoperable catalog for data and AI, supporting multi-format tables, unstructured data, and AI assets. It offers plugin support for extensibility and interoperates with Delta Sharing protocol. The catalog is fully open with OpenAPI spec and OSS implementation, providing unified governance for data and AI with asset-level access control enforced through REST APIs.
aides-jeunes
The user interface (and the main server) of the simulator of aids and social benefits for young people. It is based on the free socio-fiscal simulator Openfisca.
holohub
Holohub is a central repository for the NVIDIA Holoscan AI sensor processing community to share reference applications, operators, tutorials, and benchmarks. It includes example applications, community components, package configurations, and tutorials. Users and developers of the Holoscan platform are invited to reuse and contribute to this repository. The repository provides detailed instructions on prerequisites, building, running applications, contributing, and glossary terms. It also offers a searchable catalog of available components on the Holoscan SDK User Guide website.
qrev
QRev is an open-source alternative to Salesforce, offering AI agents to scale sales organizations infinitely. It aims to provide digital workers for various sales roles or a superagent named Qai. The tech stack includes TypeScript for frontend, NodeJS for backend, MongoDB for app server database, ChromaDB for vector database, SQLite for AI server SQL relational database, and Langchain for LLM tooling. The tool allows users to run client app, app server, and AI server components. It requires Node.js and MongoDB to be installed, and provides detailed setup instructions in the README file.
frontend
Nuclia frontend apps and libraries repository contains various frontend applications and libraries for the Nuclia platform. It includes components such as Dashboard, Widget, SDK, Sistema (design system), NucliaDB admin, CI/CD Deployment, and Maintenance page. The repository provides detailed instructions on installation, dependencies, and usage of these components for both Nuclia employees and external developers. It also covers deployment processes for different components and tools like ArgoCD for monitoring deployments and logs. The repository aims to facilitate the development, testing, and deployment of frontend applications within the Nuclia ecosystem.
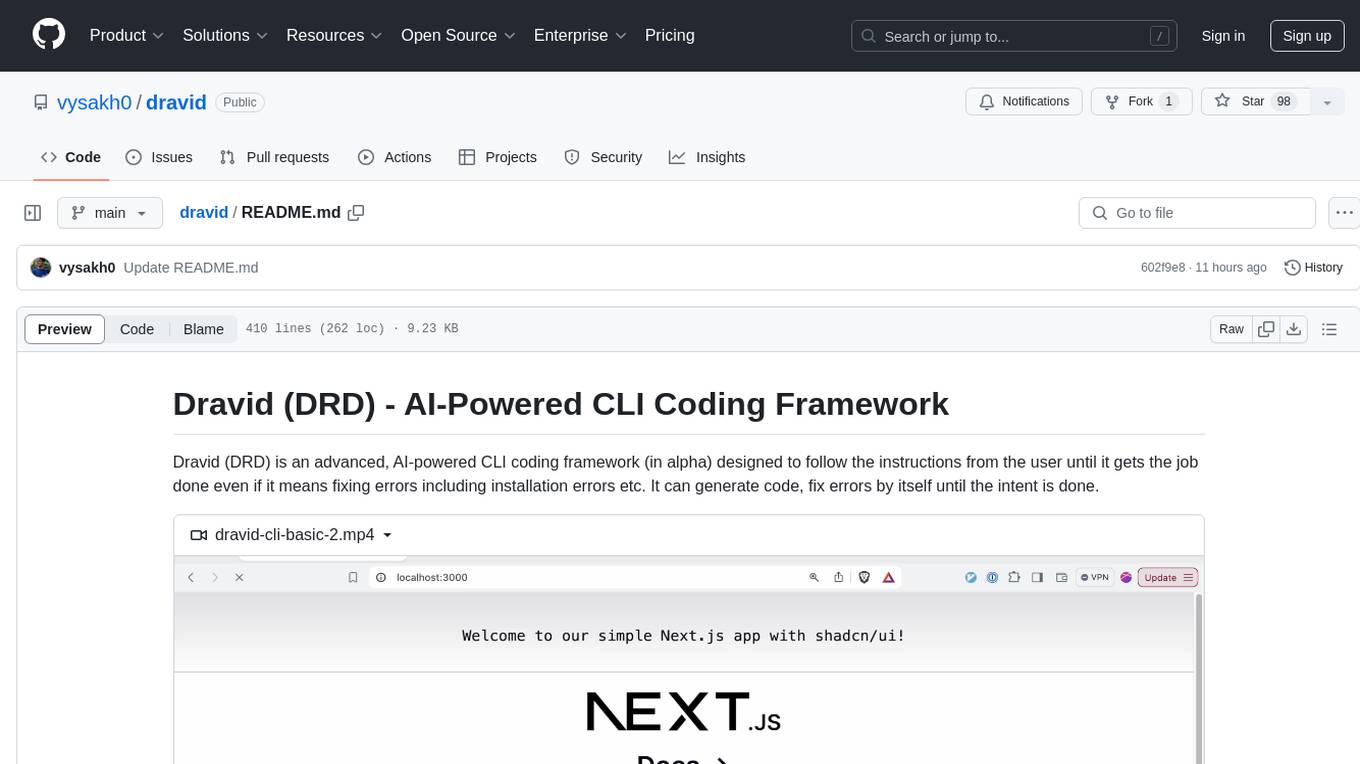
dravid
Dravid (DRD) is an advanced, AI-powered CLI coding framework designed to follow user instructions until the job is completed, including fixing errors. It can generate code, fix errors, handle image queries, manage file operations, integrate with external APIs, and provide a development server with error handling. Dravid is extensible and requires Python 3.7+ and CLAUDE_API_KEY. Users can interact with Dravid through CLI commands for various tasks like creating projects, asking questions, generating content, handling metadata, and file-specific queries. It supports use cases like Next.js project development, working with existing projects, exploring new languages, Ruby on Rails project development, and Python project development. Dravid's project structure includes directories for source code, CLI modules, API interaction, utility functions, AI prompt templates, metadata management, and tests. Contributions are welcome, and development setup involves cloning the repository, installing dependencies with Poetry, setting up environment variables, and using Dravid for project enhancements.
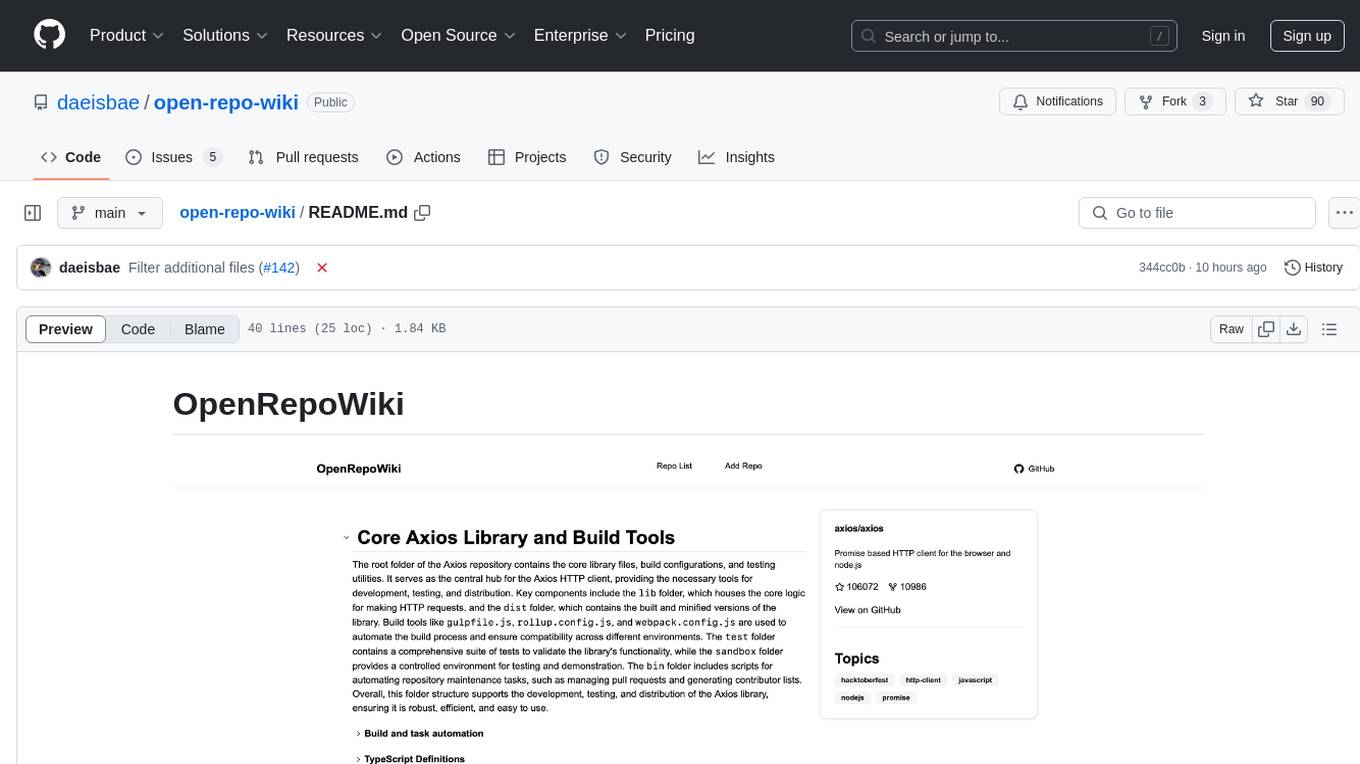
open-repo-wiki
OpenRepoWiki is a tool designed to automatically generate a comprehensive wiki page for any GitHub repository. It simplifies the process of understanding the purpose, functionality, and core components of a repository by analyzing its code structure, identifying key files and functions, and providing explanations. The tool aims to assist individuals who want to learn how to build various projects by providing a summarized overview of the repository's contents. OpenRepoWiki requires certain dependencies such as Google AI Studio or Deepseek API Key, PostgreSQL for storing repository information, Github API Key for accessing repository data, and Amazon S3 for optional usage. Users can configure the tool by setting up environment variables, installing dependencies, building the server, and running the application. It is recommended to consider the token usage and opt for cost-effective options when utilizing the tool.
AgentIQ
AgentIQ is a flexible library designed to seamlessly integrate enterprise agents with various data sources and tools. It enables true composability by treating agents, tools, and workflows as simple function calls. With features like framework agnosticism, reusability, rapid development, profiling, observability, evaluation system, user interface, and MCP compatibility, AgentIQ empowers developers to move quickly, experiment freely, and ensure reliability across agent-driven projects.
geti-sdk
The Intel® Geti™ SDK is a python package that enables teams to rapidly develop AI models by easing the complexities of model development and enhancing collaboration between teams. It provides tools to interact with an Intel® Geti™ server via the REST API, allowing for project creation, downloading, uploading, deploying for local inference with OpenVINO, setting project and model configuration, launching and monitoring training jobs, and media upload and prediction. The SDK also includes tutorial-style Jupyter notebooks demonstrating its usage.
grafana-llm-app
This repository contains separate packages for Grafana LLM Plugin and the @grafana/llm package for interfacing with it. The packages are tightly coupled and developed together with identical dependencies. The repository provides instructions for developing the packages, including backend and frontend development, testing, and release processes.
hugescm
HugeSCM is a cloud-based version control system designed to address R&D repository size issues. It effectively manages large repositories and individual large files by separating data storage and utilizing advanced algorithms and data structures. It aims for optimal performance in handling version control operations of large-scale repositories, making it suitable for single large library R&D, AI model development, and game or driver development.
todoist-ai
Library for connecting AI agents to Todoist, enabling them to access and modify a Todoist account on the user's behalf. Tools can be used through an MCP server or integrated into other projects for AI conversational interfaces. Reusable tools allow for complete workflows, balancing flexibility and efficiency for LLMs. Early-stage project with more tools planned. Designed to provide a small set of tools for various AI interfaces.
For similar tasks

aioli
Aioli is a library for running genomics command-line tools in the browser using WebAssembly. It creates a single WebWorker to run all WebAssembly tools, shares a filesystem across modules, and efficiently mounts local files. The tool encapsulates each module for loading, does WebAssembly feature detection, and communicates with the WebWorker using the Comlink library. Users can deploy new releases and versions, and benefit from code reuse by porting existing C/C++/Rust/etc tools to WebAssembly for browser use.
AliceVision
AliceVision is a photogrammetric computer vision framework which provides a 3D reconstruction pipeline. It is designed to process images from different viewpoints and create detailed 3D models of objects or scenes. The framework includes various algorithms for feature detection, matching, and structure from motion. AliceVision is suitable for researchers, developers, and enthusiasts interested in computer vision, photogrammetry, and 3D modeling. It can be used for applications such as creating 3D models of buildings, archaeological sites, or objects for virtual reality and augmented reality experiences.
For similar jobs

NoLabs
NoLabs is an open-source biolab that provides easy access to state-of-the-art models for bio research. It supports various tasks, including drug discovery, protein analysis, and small molecule design. NoLabs aims to accelerate bio research by making inference models accessible to everyone.

OpenCRISPR
OpenCRISPR is a set of free and open gene editing systems designed by Profluent Bio. The OpenCRISPR-1 protein maintains the prototypical architecture of a Type II Cas9 nuclease but is hundreds of mutations away from SpCas9 or any other known natural CRISPR-associated protein. You can view OpenCRISPR-1 as a drop-in replacement for many protocols that need a cas9-like protein with an NGG PAM and you can even use it with canonical SpCas9 gRNAs. OpenCRISPR-1 can be fused in a deactivated or nickase format for next generation gene editing techniques like base, prime, or epigenome editing.
ersilia
The Ersilia Model Hub is a unified platform of pre-trained AI/ML models dedicated to infectious and neglected disease research. It offers an open-source, low-code solution that provides seamless access to AI/ML models for drug discovery. Models housed in the hub come from two sources: published models from literature (with due third-party acknowledgment) and custom models developed by the Ersilia team or contributors.
ontogpt
OntoGPT is a Python package for extracting structured information from text using large language models, instruction prompts, and ontology-based grounding. It provides a command line interface and a minimal web app for easy usage. The tool has been evaluated on test data and is used in related projects like TALISMAN for gene set analysis. OntoGPT enables users to extract information from text by specifying relevant terms and provides the extracted objects as output.
bia-bob
BIA `bob` is a Jupyter-based assistant for interacting with data using large language models to generate Python code. It can utilize OpenAI's chatGPT, Google's Gemini, Helmholtz' blablador, and Ollama. Users need respective accounts to access these services. Bob can assist in code generation, bug fixing, code documentation, GPU-acceleration, and offers a no-code custom Jupyter Kernel. It provides example notebooks for various tasks like bio-image analysis, model selection, and bug fixing. Installation is recommended via conda/mamba environment. Custom endpoints like blablador and ollama can be used. Google Cloud AI API integration is also supported. The tool is extensible for Python libraries to enhance Bob's functionality.

Scientific-LLM-Survey
Scientific Large Language Models (Sci-LLMs) is a repository that collects papers on scientific large language models, focusing on biology and chemistry domains. It includes textual, molecular, protein, and genomic languages, as well as multimodal language. The repository covers various large language models for tasks such as molecule property prediction, interaction prediction, protein sequence representation, protein sequence generation/design, DNA-protein interaction prediction, and RNA prediction. It also provides datasets and benchmarks for evaluating these models. The repository aims to facilitate research and development in the field of scientific language modeling.
polaris
Polaris establishes a novel, industry‑certified standard to foster the development of impactful methods in AI-based drug discovery. This library is a Python client to interact with the Polaris Hub. It allows you to download Polaris datasets and benchmarks, evaluate a custom method against a Polaris benchmark, and create and upload new datasets and benchmarks.
awesome-AI4MolConformation-MD
The 'awesome-AI4MolConformation-MD' repository focuses on protein conformations and molecular dynamics using generative artificial intelligence and deep learning. It provides resources, reviews, datasets, packages, and tools related to AI-driven molecular dynamics simulations. The repository covers a wide range of topics such as neural networks potentials, force fields, AI engines/frameworks, trajectory analysis, visualization tools, and various AI-based models for protein conformational sampling. It serves as a comprehensive guide for researchers and practitioners interested in leveraging AI for studying molecular structures and dynamics.
