
ctakes
Apache cTAKES is a Natural Language Processing (NLP) platform for clinical text.
Stars: 99
Apache cTAKES is a clinical Text Analysis and Knowledge Extraction System that focuses on extracting knowledge from clinical text through Natural Language Processing (NLP) techniques. It is modular and employs rule-based and machine learning methods to extract concepts such as symptoms, procedures, diagnoses, medications, and anatomy with attributes and standard codes. cTAKES can identify temporal events, dates, and times, placing events in a patient timeline. It supports various biomedical text processing tasks and can handle different types of clinical and health-related narratives using multiple data standards. cTAKES is widely used in research initiatives and encourages contributions from professionals, researchers, doctors, and students from diverse backgrounds.
README:
The Apache™ clinical Text Analysis and Knowledge Extraction System (cTAKES™) focuses on extracting knowledge from clinical text through Natural Language Processing (NLP) techniques.
cTAKES is engineered in a modular fashion and employs leading-edge rule-based and machine learning methods.
cTAKES has standard features for biomedical text processing software, including the ability to extract concepts such as symptoms, procedures, diagnoses, medications and anatomy with attributes and standard codes.
More powerful components can perform tasks as complex as identifying temporal events, dates and times – resulting in placement of events in a patient timeline.
Components are trained on gold standards from the biomedical as well as the general domain. This affords usability across different types of clinical narrative (e.g. radiology reports, clinical notes, discharge summaries) in various institution formats as well as other types of health-related narrative (e.g. twitter feeds), using multiple data standards (e.g. Health Level 7 (HL7), Clinical Document Architecture (CDA), Fast Healthcare Interoperability Resources (FHIR), SNOMED-CT, RxNORM).
cTAKES is the NLP platform for many initiatives across the world covering a variety of research purposes and large datasets. Contributors include professionals at medical and commercial institutions, NLP and Machine Learning researchers, Medical Doctors, and students of many disciplines and levels. We encourage people from all backgrounds to get involved! (link)
- Java 17 is required to run cTAKES 6.0.0 and higher. Java 8 or Java 11 is required to run cTAKES 5. Run this command to check your Java version:
java -version
- A license for the National Library of Medicine's Unified Medical Language System (UMLS) is required to use the named entity recognition module (dictionary lookup) with the default dictionary.
- Python 3 is required to use cTAKES Python Bridge to Java (PBJ). Run this to command to check your Python version:
python -V
[!NOTE] If you are using an integrated development environment (IDE), please see its documentation on using git, Java, Python, and Apache Maven. You should be able to use features in your IDE instead of running commands in a terminal.
### For developers:
- Apache Maven 3 is required to build cTAKES. Run this to command to check your Maven version:
mvn -version
For access to all cTAKES capabilities, download a pre-built copy of a cTAKES installation from the release area.
The names of pre-built installations follow the format apache-ctakes-#.#.#-bin.zip.
After unzipping the release file and obtaining a UMLS license, use the UMLS Package Fetcher GUI to install a copy of the
default dictionary for Named Entity Recognition (NER) using cTAKES Fast Dictionary Lookup.
You can then use the Piper File Submitter GUI to submit jobs,
or run any of the scripts in the bin/ directory.
All source code for cTAKES versions 5+ is available from the cTAKES GitHub repository.
- Clone the cTAKES code repository using git.
git clone https://github.com/apache/ctakes.git
- Compile the cTAKES code using Apache Maven. In your cTAKES root directory, run this command:
mvn clean compile
- Download the default cTAKES dictionary zip file.
- Copy the contents of the zip file to the
resources/org/apache/ctakes/dictionary/lookup/fastdirectory.
[!TIP] As an alternative to steps 3 and 4, you can use the UMLS Package Fetcher GUI. Run the class
DictionaryDownloader.javato launch that tool, or use thegetUmlsDictionaryscript if using a full build of cTAKES.
- Run the cTAKES default pipeline using the Java class
PiperFileRunner.java. To use the Piper File Submitter GUI, run thePiperRunnerGui.javaclass.
[!NOTE] To run the cTAKES Java classes, the full Java classpath must be configured. Setting up a classpath is beyond the scope of this document.
An integrated development environment (IDE) should set up the classpath for you, please see its documentation.
[!IMPORTANT] You cannot run scripts in the
bin/directory within a development environment. Within a cTAKES development environment you can run Java classes and Maven profiles, but no scripts in thebindirectory.
[!TIP] You can build your own cTAKES installation from a development environment using Apache Maven. A cTAKES installation is required to run scripts in the
bin/directory.
- Build using Apache Maven:
mvn clean compile package
[!NOTE] If you are using an integrated development environment (IDE), please see its documentation on using Apache Maven.
After packaging, there should be tar and zip files for apache-ctakes-#.#.#.-bin and apache-ctakes-#.#.#.-src in your ctakes-distribution/target/ directory.
7. Unzip the apache-ctakes-#.#.#.-bin into a directory outside your cTAKES development area.
You can write to the cTAKES user and developer mailing lists: user at ctakes.apache.org and dev at apache.ctakes.org
and find answers to previously asked questions by searching the user
and developer mail archives.
For Tasks:
Click tags to check more tools for each tasksFor Jobs:
Alternative AI tools for ctakes
Similar Open Source Tools

ctakes
Apache cTAKES is a clinical Text Analysis and Knowledge Extraction System that focuses on extracting knowledge from clinical text through Natural Language Processing (NLP) techniques. It is modular and employs rule-based and machine learning methods to extract concepts such as symptoms, procedures, diagnoses, medications, and anatomy with attributes and standard codes. cTAKES can identify temporal events, dates, and times, placing events in a patient timeline. It supports various biomedical text processing tasks and can handle different types of clinical and health-related narratives using multiple data standards. cTAKES is widely used in research initiatives and encourages contributions from professionals, researchers, doctors, and students from diverse backgrounds.
jaison-core
J.A.I.son is a Python project designed for generating responses using various components and applications. It requires specific plugins like STT, T2T, TTSG, and TTSC to function properly. Users can customize responses, voice, and configurations. The project provides a Discord bot, Twitch events and chat integration, and VTube Studio Animation Hotkeyer. It also offers features for managing conversation history, training AI models, and monitoring conversations.
council
Council is an open-source platform designed for the rapid development and deployment of customized generative AI applications using teams of agents. It extends the LLM tool ecosystem by providing advanced control flow and scalable oversight for AI agents. Users can create sophisticated agents with predictable behavior by leveraging Council's powerful approach to control flow using Controllers, Filters, Evaluators, and Budgets. The framework allows for automated routing between agents, comparing, evaluating, and selecting the best results for a task. Council aims to facilitate packaging and deploying agents at scale on multiple platforms while enabling enterprise-grade monitoring and quality control.

guidellm
GuideLLM is a platform for evaluating and optimizing the deployment of large language models (LLMs). By simulating real-world inference workloads, GuideLLM enables users to assess the performance, resource requirements, and cost implications of deploying LLMs on various hardware configurations. This approach ensures efficient, scalable, and cost-effective LLM inference serving while maintaining high service quality. The tool provides features for performance evaluation, resource optimization, cost estimation, and scalability testing.
codebase-context-spec
The Codebase Context Specification (CCS) project aims to standardize embedding contextual information within codebases to enhance understanding for both AI and human developers. It introduces a convention similar to `.env` and `.editorconfig` files but focused on documenting code for both AI and humans. By providing structured contextual metadata, collaborative documentation guidelines, and standardized context files, developers can improve code comprehension, collaboration, and development efficiency. The project includes a linter for validating context files and provides guidelines for using the specification with AI assistants. Tooling recommendations suggest creating memory systems, IDE plugins, AI model integrations, and agents for context creation and utilization. Future directions include integration with existing documentation systems, dynamic context generation, and support for explicit context overriding.
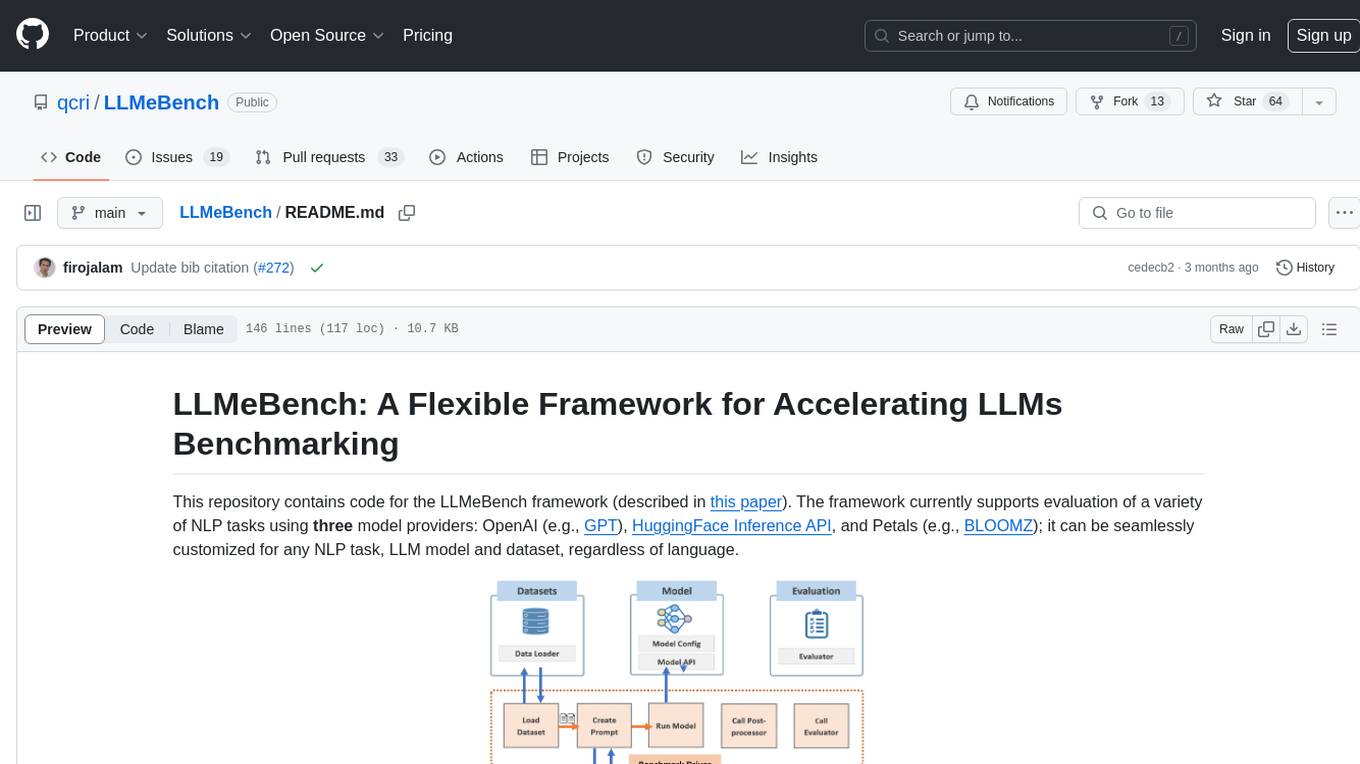
LLMeBench
LLMeBench is a flexible framework designed for accelerating benchmarking of Large Language Models (LLMs) in the field of Natural Language Processing (NLP). It supports evaluation of various NLP tasks using model providers like OpenAI, HuggingFace Inference API, and Petals. The framework is customizable for different NLP tasks, LLM models, and datasets across multiple languages. It features extensive caching capabilities, supports zero- and few-shot learning paradigms, and allows on-the-fly dataset download and caching. LLMeBench is open-source and continuously expanding to support new models accessible through APIs.
CoLLM
CoLLM is a novel method that integrates collaborative information into Large Language Models (LLMs) for recommendation. It converts recommendation data into language prompts, encodes them with both textual and collaborative information, and uses a two-step tuning method to train the model. The method incorporates user/item ID fields in prompts and employs a conventional collaborative model to generate user/item representations. CoLLM is built upon MiniGPT-4 and utilizes pretrained Vicuna weights for training.
dlio_benchmark
DLIO is an I/O benchmark tool designed for Deep Learning applications. It emulates modern deep learning applications using Benchmark Runner, Data Generator, Format Handler, and I/O Profiler modules. Users can configure various I/O patterns, data loaders, data formats, datasets, and parameters. The tool is aimed at emulating the I/O behavior of deep learning applications and provides a modular design for flexibility and customization.
aici
The Artificial Intelligence Controller Interface (AICI) lets you build Controllers that constrain and direct output of a Large Language Model (LLM) in real time. Controllers are flexible programs capable of implementing constrained decoding, dynamic editing of prompts and generated text, and coordinating execution across multiple, parallel generations. Controllers incorporate custom logic during the token-by-token decoding and maintain state during an LLM request. This allows diverse Controller strategies, from programmatic or query-based decoding to multi-agent conversations to execute efficiently in tight integration with the LLM itself.
aiid
The Artificial Intelligence Incident Database (AIID) is a collection of incidents involving the development and use of artificial intelligence (AI). The database is designed to help researchers, policymakers, and the public understand the potential risks and benefits of AI, and to inform the development of policies and practices to mitigate the risks and promote the benefits of AI. The AIID is a collaborative project involving researchers from the University of California, Berkeley, the University of Washington, and the University of Toronto.
easydist
EasyDist is an automated parallelization system and infrastructure designed for multiple ecosystems. It offers usability by making parallelizing training or inference code effortless with just a single line of change. It ensures ecological compatibility by serving as a centralized source of truth for SPMD rules at the operator-level for various machine learning frameworks. EasyDist decouples auto-parallel algorithms from specific frameworks and IRs, allowing for the development and benchmarking of different auto-parallel algorithms in a flexible manner. The architecture includes MetaOp, MetaIR, and the ShardCombine Algorithm for SPMD sharding rules without manual annotations.
comfyui_LLM_party
COMFYUI LLM PARTY is a node library designed for LLM workflow development in ComfyUI, an extremely minimalist UI interface primarily used for AI drawing and SD model-based workflows. The project aims to provide a complete set of nodes for constructing LLM workflows, enabling users to easily integrate them into existing SD workflows. It features various functionalities such as API integration, local large model integration, RAG support, code interpreters, online queries, conditional statements, looping links for large models, persona mask attachment, and tool invocations for weather lookup, time lookup, knowledge base, code execution, web search, and single-page search. Users can rapidly develop web applications using API + Streamlit and utilize LLM as a tool node. Additionally, the project includes an omnipotent interpreter node that allows the large model to perform any task, with recommendations to use the 'show_text' node for display output.
craftium
Craftium is an open-source platform based on the Minetest voxel game engine and the Gymnasium and PettingZoo APIs, designed for creating fast, rich, and diverse single and multi-agent environments. It allows for connecting to Craftium's Python process, executing actions as keyboard and mouse controls, extending the Lua API for creating RL environments and tasks, and supporting client/server synchronization for slow agents. Craftium is fully extensible, extensively documented, modern RL API compatible, fully open source, and eliminates the need for Java. It offers a variety of environments for research and development in reinforcement learning.
agentok
Agentok Studio is a visual tool built for AutoGen, a cutting-edge agent framework from Microsoft and various contributors. It offers intuitive visual tools to simplify the construction and management of complex agent-based workflows. Users can create workflows visually as graphs, chat with agents, and share flow templates. The tool is designed to streamline the development process for creators and developers working on next-generation Multi-Agent Applications.

verifAI
VerifAI is a document-based question-answering system that addresses hallucinations in generative large language models and search engines. It retrieves relevant documents, generates answers with references, and verifies answers for accuracy. The engine uses generative search technology and a verification model to ensure no misinformation. VerifAI supports various document formats and offers user registration with a React.js interface. It is open-source and designed to be user-friendly, making it accessible for anyone to use.
Guardrails
Guardrails is a security tool designed to help developers identify and fix security vulnerabilities in their code. It provides automated scanning and analysis of code repositories to detect potential security issues, such as sensitive data exposure, injection attacks, and insecure configurations. By integrating Guardrails into the development workflow, teams can proactively address security concerns and reduce the risk of security breaches. The tool offers detailed reports and actionable recommendations to guide developers in remediation efforts, ultimately improving the overall security posture of the codebase. Guardrails supports multiple programming languages and frameworks, making it versatile and adaptable to different development environments. With its user-friendly interface and seamless integration with popular version control systems, Guardrails empowers developers to prioritize security without compromising productivity.
For similar tasks

ctakes
Apache cTAKES is a clinical Text Analysis and Knowledge Extraction System that focuses on extracting knowledge from clinical text through Natural Language Processing (NLP) techniques. It is modular and employs rule-based and machine learning methods to extract concepts such as symptoms, procedures, diagnoses, medications, and anatomy with attributes and standard codes. cTAKES can identify temporal events, dates, and times, placing events in a patient timeline. It supports various biomedical text processing tasks and can handle different types of clinical and health-related narratives using multiple data standards. cTAKES is widely used in research initiatives and encourages contributions from professionals, researchers, doctors, and students from diverse backgrounds.
For similar jobs

ctakes
Apache cTAKES is a clinical Text Analysis and Knowledge Extraction System that focuses on extracting knowledge from clinical text through Natural Language Processing (NLP) techniques. It is modular and employs rule-based and machine learning methods to extract concepts such as symptoms, procedures, diagnoses, medications, and anatomy with attributes and standard codes. cTAKES can identify temporal events, dates, and times, placing events in a patient timeline. It supports various biomedical text processing tasks and can handle different types of clinical and health-related narratives using multiple data standards. cTAKES is widely used in research initiatives and encourages contributions from professionals, researchers, doctors, and students from diverse backgrounds.
seismometer
Seismometer is a suite of tools designed to evaluate AI model performance in healthcare settings. It helps healthcare organizations assess the accuracy of AI models and ensure equitable care for diverse patient populations. The tool allows users to validate model performance using standardized evaluation criteria based on local data and workflows. It includes templates for analyzing statistical performance, fairness across different cohorts, and the impact of interventions on outcomes. Seismometer is continuously evolving to incorporate new validation and analysis techniques.
fuse-med-ml
FuseMedML is a Python framework designed to accelerate machine learning-based discovery in the medical field by promoting code reuse. It provides a flexible design concept where data is stored in a nested dictionary, allowing easy handling of multi-modality information. The framework includes components for creating custom models, loss functions, metrics, and data processing operators. Additionally, FuseMedML offers 'batteries included' key components such as fuse.data for data processing, fuse.eval for model evaluation, and fuse.dl for reusable deep learning components. It supports PyTorch and PyTorch Lightning libraries and encourages the creation of domain extensions for specific medical domains.
MedLLMsPracticalGuide
This repository serves as a practical guide for Medical Large Language Models (Medical LLMs) and provides resources, surveys, and tools for building, fine-tuning, and utilizing LLMs in the medical domain. It covers a wide range of topics including pre-training, fine-tuning, downstream biomedical tasks, clinical applications, challenges, future directions, and more. The repository aims to provide insights into the opportunities and challenges of LLMs in medicine and serve as a practical resource for constructing effective medical LLMs.
hi-ml
The Microsoft Health Intelligence Machine Learning Toolbox is a repository that provides low-level and high-level building blocks for Machine Learning / AI researchers and practitioners. It simplifies and streamlines work on deep learning models for healthcare and life sciences by offering tested components such as data loaders, pre-processing tools, deep learning models, and cloud integration utilities. The repository includes two Python packages, 'hi-ml-azure' for helper functions in AzureML, 'hi-ml' for ML components, and 'hi-ml-cpath' for models and workflows related to histopathology images.
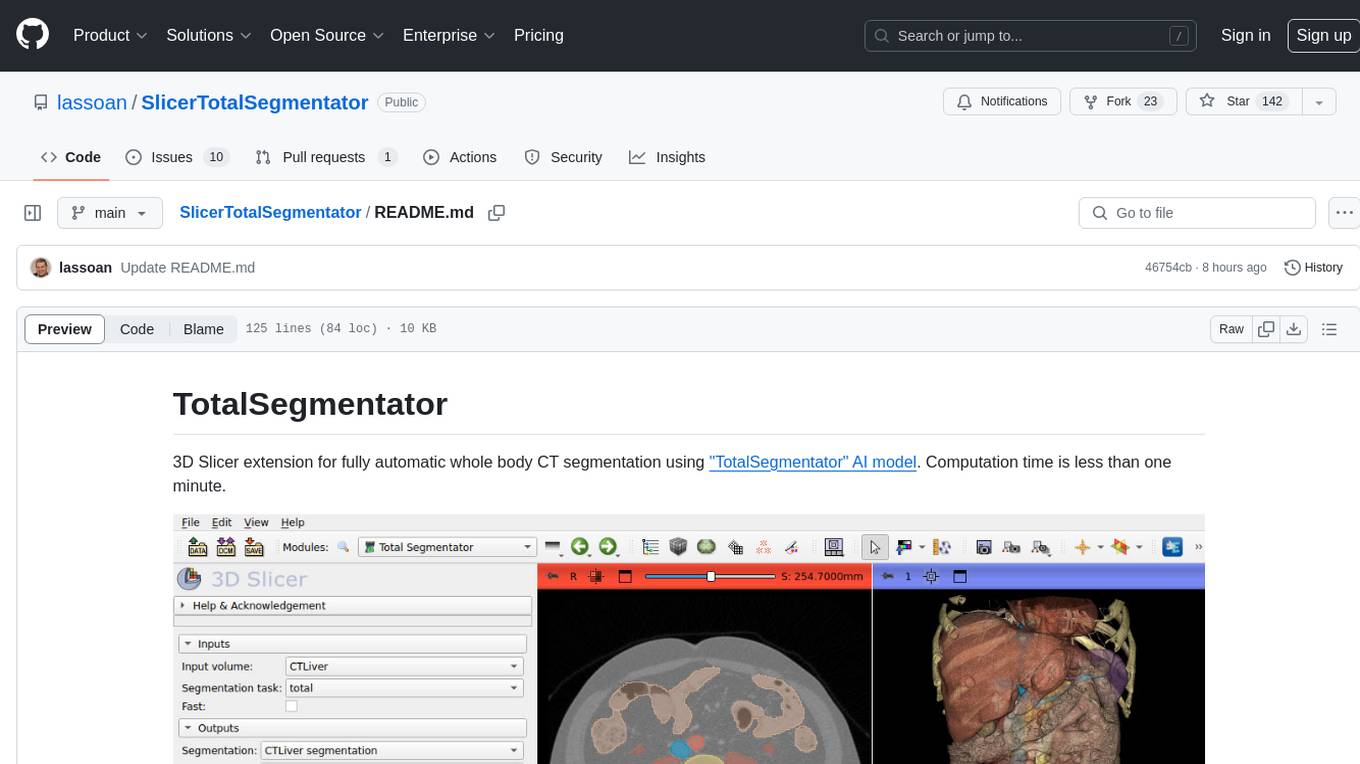
SlicerTotalSegmentator
TotalSegmentator is a 3D Slicer extension designed for fully automatic whole body CT segmentation using the 'TotalSegmentator' AI model. The computation time is less than one minute, making it efficient for research purposes. Users can set up GPU acceleration for faster segmentation. The tool provides a user-friendly interface for loading CT images, creating segmentations, and displaying results in 3D. Troubleshooting steps are available for common issues such as failed computation, GPU errors, and inaccurate segmentations. Contributions to the extension are welcome, following 3D Slicer contribution guidelines.
machine-learning-research
The 'machine-learning-research' repository is a comprehensive collection of resources related to mathematics, machine learning, deep learning, artificial intelligence, data science, and various scientific fields. It includes materials such as courses, tutorials, books, podcasts, communities, online courses, papers, and dissertations. The repository covers topics ranging from fundamental math skills to advanced machine learning concepts, with a focus on applications in healthcare, genetics, computational biology, precision health, and AI in science. It serves as a valuable resource for individuals interested in learning and researching in the fields of machine learning and related disciplines.
LLMonFHIR
LLMonFHIR is an iOS application that utilizes large language models (LLMs) to interpret and provide context around patient data in the Fast Healthcare Interoperability Resources (FHIR) format. It connects to the OpenAI GPT API to analyze FHIR resources, supports multiple languages, and allows users to interact with their health data stored in the Apple Health app. The app aims to simplify complex health records, provide insights, and facilitate deeper understanding through a conversational interface. However, it is an experimental app for informational purposes only and should not be used as a substitute for professional medical advice. Users are advised to verify information provided by AI models and consult healthcare professionals for personalized advice.