
aigt
Deep learning software modules for image-guided medical procedures
Stars: 59
AIGT is a repository containing scripts for deep learning in guided medical interventions, focusing on ultrasound imaging. It provides a complete workflow from formatting and annotations to real-time model deployment. Users can set up an Anaconda environment, run Slicer notebooks, acquire tracked ultrasound data, and process exported data for training. The repository includes tools for segmentation, image export, and annotation creation.
README:
This repository contains scripts for deep learning for guided medical interventions. For some projects, the complete workflow is implemented, from formatting and annotations to deployment of models in real time. Most projects use ultrasound imaging.
- Install a recent Anaconda version for Python 3.9 version from the Anaconda download page
- Clone this repository on your computer. Your local clone may be in a path like
c:\dev\aigt - Start the Anaconda Prompt application and navigate to the environment setup folder
cd c:\dev\aigt\SetupAnaconda - Run the setup_env.bat file to create environment in a folder, e.g.
setup_env.bat c:\dev\dlenvThis will install TensorFlow 2.0 and other packages that are used by projects. The previous environment setup script (for TensorFlow v1 is still available assetup_env_tf1.bat
Please do not commit/push these local files, as everybody sets them up with values that only apply to their environment.
-
local_vars.py - Some notebooks require a file in the Notebooks folder of your local repository clone, named local_vars.py. This file should define the root_folder variable. The file may just contain this single line of code:
root_folder = r"c:\Data". - girder_apikey_read.py - Some notebooks require a file named girder_apikey_read.py with a single line of code that specifies your personal API key to the private data collection. If you work with non-public data stored on a Girder server, ask your supervisor for a Girder account and how to generate API keys for yourself.
- Install Slicer 4.11 or newer version (later than 2019-09-16 is recommended, for full functionality)
- Install the SlicerJupyter extension for Slicer, and follow the extension user guide to add Slicer as a kernel in Jupyter Notebook (use the Copy command to clipboard button and paste it in the active Anaconda environment).
- If you have a GPU and would like Slicer's TensorFlow to use it, then install CUDA 10.1 and cuDNN 7.6.5. GPUs can make training of models much faster, but may not significantly speed up trained models for prediction compared to CPUs.
- Some users have reported that they needed to install Visual Studio (2015 or later) to be able to use TensorFlow.
- Install additional packages in the Slicer python environment, to be able to run all Slicer notebooks. Use the Python console of Slicer to run this command:
pip_install("tensorflow opencv-contrib-python girder_client pandas nbformat nbconvert")
- To run notebooks, start the Anaconda command prompt, navigate to the aigt folder, and type the
jupyter notebookcommand.
- Use the Sequences extension in 3D Slicer to record tracked ultrasound sequences.
- Record Image_Image (the ultrasound in the Image coordinate system) and ImageToReference transform sequences. Note that Slicer cannot record transformed images, so recording Image_Reference is not an option.
- If you work on segmentation, you can use the SingleSliceSegmentation Slicer module in this repository to speed up manual segmentation.
- Create annotations by placing fiducials or creating segmentations with the SingleSliceSegmentation module.
- For segmentations, you may use the SingleSliceSegmentation module to export images and segmentations in png file format.
- Use scripts in the Notebooks/Slicer folder to export images and annotations.
- It is recommended that you save separate sequences for validation and testing
- Use Notebooks/FoldersToSavedArrays to save data sequences as single files for faster data loading during training.
 This work was supported through CANARIE’s Research Software Program through RS3-036. Principal investigator: Gabor Fichtinger.
This work was supported through CANARIE’s Research Software Program through RS3-036. Principal investigator: Gabor Fichtinger.
For Tasks:
Click tags to check more tools for each tasksFor Jobs:
Alternative AI tools for aigt
Similar Open Source Tools

aigt
AIGT is a repository containing scripts for deep learning in guided medical interventions, focusing on ultrasound imaging. It provides a complete workflow from formatting and annotations to real-time model deployment. Users can set up an Anaconda environment, run Slicer notebooks, acquire tracked ultrasound data, and process exported data for training. The repository includes tools for segmentation, image export, and annotation creation.
aiCoder
aiCoder is an AI-powered tool designed to streamline the coding process by automating repetitive tasks, providing intelligent code suggestions, and facilitating the integration of new features into existing codebases. It offers a chat interface for natural language interactions, methods and stubs lists for code modification, and settings customization for project-specific prompts. Users can leverage aiCoder to enhance code quality, focus on higher-level design, and save time during development.
Open_Data_QnA
Open Data QnA is a Python library that allows users to interact with their PostgreSQL or BigQuery databases in a conversational manner, without needing to write SQL queries. The library leverages Large Language Models (LLMs) to bridge the gap between human language and database queries, enabling users to ask questions in natural language and receive informative responses. It offers features such as conversational querying with multiturn support, table grouping, multi schema/dataset support, SQL generation, query refinement, natural language responses, visualizations, and extensibility. The library is built on a modular design and supports various components like Database Connectors, Vector Stores, and Agents for SQL generation, validation, debugging, descriptions, embeddings, responses, and visualizations.
aici
The Artificial Intelligence Controller Interface (AICI) lets you build Controllers that constrain and direct output of a Large Language Model (LLM) in real time. Controllers are flexible programs capable of implementing constrained decoding, dynamic editing of prompts and generated text, and coordinating execution across multiple, parallel generations. Controllers incorporate custom logic during the token-by-token decoding and maintain state during an LLM request. This allows diverse Controller strategies, from programmatic or query-based decoding to multi-agent conversations to execute efficiently in tight integration with the LLM itself.
jaison-core
J.A.I.son is a Python project designed for generating responses using various components and applications. It requires specific plugins like STT, T2T, TTSG, and TTSC to function properly. Users can customize responses, voice, and configurations. The project provides a Discord bot, Twitch events and chat integration, and VTube Studio Animation Hotkeyer. It also offers features for managing conversation history, training AI models, and monitoring conversations.
vespa
Vespa is a platform that performs operations such as selecting a subset of data in a large corpus, evaluating machine-learned models over the selected data, organizing and aggregating it, and returning it, typically in less than 100 milliseconds, all while the data corpus is continuously changing. It has been in development for many years and is used on a number of large internet services and apps which serve hundreds of thousands of queries from Vespa per second.

airbroke
Airbroke is an open-source error catcher tool designed for modern web applications. It provides a PostgreSQL-based backend with an Airbrake-compatible HTTP collector endpoint and a React-based frontend for error management. The tool focuses on simplicity, maintaining a small database footprint even under heavy data ingestion. Users can ask AI about issues, replay HTTP exceptions, and save/manage bookmarks for important occurrences. Airbroke supports multiple OAuth providers for secure user authentication and offers occurrence charts for better insights into error occurrences. The tool can be deployed in various ways, including building from source, using Docker images, deploying on Vercel, Render.com, Kubernetes with Helm, or Docker Compose. It requires Node.js, PostgreSQL, and specific system resources for deployment.

raggenie
RAGGENIE is a low-code RAG builder tool designed to simplify the creation of conversational AI applications. It offers out-of-the-box plugins for connecting to various data sources and building conversational AI on top of them, including integration with pre-built agents for actions. The tool is open-source under the MIT license, with a current focus on making it easy to build RAG applications and future plans for maintenance, monitoring, and transitioning applications from pilots to production.
ultimate-rvc
Ultimate RVC is an extension of AiCoverGen, offering new features and improvements for generating audio content using RVC. It is designed for users looking to integrate singing functionality into AI assistants/chatbots/vtubers, create character voices for songs or books, and train voice models. The tool provides easy setup, voice conversion enhancements, TTS functionality, voice model training suite, caching system, UI improvements, and support for custom configurations. It is available for local and Google Colab use, with a PyPI package for easy access. The tool also offers CLI usage and customization through environment variables.

project_alice
Alice is an agentic workflow framework that integrates task execution and intelligent chat capabilities. It provides a flexible environment for creating, managing, and deploying AI agents for various purposes, leveraging a microservices architecture with MongoDB for data persistence. The framework consists of components like APIs, agents, tasks, and chats that interact to produce outputs through files, messages, task results, and URL references. Users can create, test, and deploy agentic solutions in a human-language framework, making it easy to engage with by both users and agents. The tool offers an open-source option, user management, flexible model deployment, and programmatic access to tasks and chats.
unstructured
The `unstructured` library provides open-source components for ingesting and pre-processing images and text documents, such as PDFs, HTML, Word docs, and many more. The use cases of `unstructured` revolve around streamlining and optimizing the data processing workflow for LLMs. `unstructured` modular functions and connectors form a cohesive system that simplifies data ingestion and pre-processing, making it adaptable to different platforms and efficient in transforming unstructured data into structured outputs.
lumigator
Lumigator is an open-source platform developed by Mozilla.ai to help users select the most suitable language model for their specific needs. It supports the evaluation of summarization tasks using sequence-to-sequence models such as BART and BERT, as well as causal models like GPT and Mistral. The platform aims to make model selection transparent, efficient, and empowering by providing a framework for comparing LLMs using task-specific metrics to evaluate how well a model fits a project's needs. Lumigator is in the early stages of development and plans to expand support to additional machine learning tasks and use cases in the future.
nx_open
The `nx_open` repository contains open-source components for the Network Optix Meta Platform, used to build products like Nx Witness Video Management System. It includes source code, specifications, and a Desktop Client. The repository is licensed under Mozilla Public License 2.0. Users can build the Desktop Client and customize it using a zip file. The build environment supports Windows, Linux, and macOS platforms with specific prerequisites. The repository provides scripts for building, signing executable files, and running the Desktop Client. Compatibility with VMS Server versions is crucial, and automatic VMS updates are disabled for the open-source Desktop Client.
serverless-pdf-chat
The serverless-pdf-chat repository contains a sample application that allows users to ask natural language questions of any PDF document they upload. It leverages serverless services like Amazon Bedrock, AWS Lambda, and Amazon DynamoDB to provide text generation and analysis capabilities. The application architecture involves uploading a PDF document to an S3 bucket, extracting metadata, converting text to vectors, and using a LangChain to search for information related to user prompts. The application is not intended for production use and serves as a demonstration and educational tool.
azure-search-openai-demo
This sample demonstrates a few approaches for creating ChatGPT-like experiences over your own data using the Retrieval Augmented Generation pattern. It uses Azure OpenAI Service to access a GPT model (gpt-35-turbo), and Azure AI Search for data indexing and retrieval. The repo includes sample data so it's ready to try end to end. In this sample application we use a fictitious company called Contoso Electronics, and the experience allows its employees to ask questions about the benefits, internal policies, as well as job descriptions and roles.
REINVENT4
REINVENT is a molecular design tool for de novo design, scaffold hopping, R-group replacement, linker design, molecule optimization, and other small molecule design tasks. It uses a Reinforcement Learning (RL) algorithm to generate optimized molecules compliant with a user-defined property profile defined as a multi-component score. Transfer Learning (TL) can be used to create or pre-train a model that generates molecules closer to a set of input molecules.
For similar tasks

aigt
AIGT is a repository containing scripts for deep learning in guided medical interventions, focusing on ultrasound imaging. It provides a complete workflow from formatting and annotations to real-time model deployment. Users can set up an Anaconda environment, run Slicer notebooks, acquire tracked ultrasound data, and process exported data for training. The repository includes tools for segmentation, image export, and annotation creation.
cursor-talk-to-figma-mcp
This project implements a Model Context Protocol (MCP) integration between Cursor AI and Figma, allowing Cursor to communicate with Figma for reading designs and modifying them programmatically. It provides tools for interacting with Figma such as creating elements, modifying text content, styling, layout & organization, components & styles, export & advanced features, and connection management. The project structure includes a TypeScript MCP server for Figma integration, a Figma plugin for communicating with Cursor, and a WebSocket server for facilitating communication between the MCP server and Figma plugin.
aistore
AIStore is a lightweight object storage system designed for AI applications. It is highly scalable, reliable, and easy to use. AIStore can be deployed on any commodity hardware, and it can be used to store and manage large datasets for deep learning and other AI applications.

cl-waffe2
cl-waffe2 is an experimental deep learning framework in Common Lisp, providing fast, systematic, and customizable matrix operations, reverse mode tape-based Automatic Differentiation, and neural network model building and training features accelerated by a JIT Compiler. It offers abstraction layers, extensibility, inlining, graph-level optimization, visualization, debugging, systematic nodes, and symbolic differentiation. Users can easily write extensions and optimize their networks without overheads. The framework is designed to eliminate barriers between users and developers, allowing for easy customization and extension.
DeepLearing-Interview-Awesome-2024
DeepLearning-Interview-Awesome-2024 is a repository that covers various topics related to deep learning, computer vision, big models (LLMs), autonomous driving, smart healthcare, and more. It provides a collection of interview questions with detailed explanations sourced from recent academic papers and industry developments. The repository is aimed at assisting individuals in academic research, work innovation, and job interviews. It includes six major modules covering topics such as large language models (LLMs), computer vision models, common problems in computer vision and perception algorithms, deep learning basics and frameworks, as well as specific tasks like 3D object detection, medical image segmentation, and more.
PINNACLE
PINNACLE is a flexible geometric deep learning approach that trains on contextualized protein interaction networks to generate context-aware protein representations. It provides protein representations split across various cell-type contexts from different tissues and organs. The tool can be fine-tuned to study the genomic effects of drugs and nominate promising protein targets and cell-type contexts for further investigation. PINNACLE exemplifies the paradigm of incorporating context-specific effects for studying biological systems, especially the impact of disease and therapeutics.
ai-hands-on
A complete, hands-on guide to becoming an AI Engineer. This repository is designed to help you learn AI from first principles, build real neural networks, and understand modern LLM systems end-to-end. Progress through math, PyTorch, deep learning, transformers, RAG, and OCR with clean, intuitive Jupyter notebooks guiding you at every step. Suitable for beginners and engineers leveling up, providing clarity, structure, and intuition to build real AI systems.
For similar jobs
grand-challenge.org
Grand Challenge is a platform that provides access to large amounts of annotated training data, objective comparisons of state-of-the-art machine learning solutions, and clinical validation using real-world data. It assists researchers, data scientists, and clinicians in collaborating to develop robust machine learning solutions to problems in biomedical imaging.
Detection-and-Classification-of-Alzheimers-Disease
This tool is designed to detect and classify Alzheimer's Disease using Deep Learning and Machine Learning algorithms on an early basis, which is further optimized using the Crow Search Algorithm (CSA). Alzheimer's is a fatal disease, and early detection is crucial for patients to predetermine their condition and prevent its progression. By analyzing MRI scanned images using Artificial Intelligence technology, this tool can classify patients who may or may not develop AD in the future. The CSA algorithm, combined with ML algorithms, has proven to be the most effective approach for this purpose.

OpenCRISPR
OpenCRISPR is a set of free and open gene editing systems designed by Profluent Bio. The OpenCRISPR-1 protein maintains the prototypical architecture of a Type II Cas9 nuclease but is hundreds of mutations away from SpCas9 or any other known natural CRISPR-associated protein. You can view OpenCRISPR-1 as a drop-in replacement for many protocols that need a cas9-like protein with an NGG PAM and you can even use it with canonical SpCas9 gRNAs. OpenCRISPR-1 can be fused in a deactivated or nickase format for next generation gene editing techniques like base, prime, or epigenome editing.

AlphaFold3
AlphaFold3 is an implementation of the Alpha Fold 3 model in PyTorch for accurate structure prediction of biomolecular interactions. It includes modules for genetic diffusion and full model examples for forward pass computations. The tool allows users to generate random pair and single representations, operate on atomic coordinates, and perform structure predictions based on input tensors. The implementation also provides functionalities for training and evaluating the model.
fuse-med-ml
FuseMedML is a Python framework designed to accelerate machine learning-based discovery in the medical field by promoting code reuse. It provides a flexible design concept where data is stored in a nested dictionary, allowing easy handling of multi-modality information. The framework includes components for creating custom models, loss functions, metrics, and data processing operators. Additionally, FuseMedML offers 'batteries included' key components such as fuse.data for data processing, fuse.eval for model evaluation, and fuse.dl for reusable deep learning components. It supports PyTorch and PyTorch Lightning libraries and encourages the creation of domain extensions for specific medical domains.
hi-ml
The Microsoft Health Intelligence Machine Learning Toolbox is a repository that provides low-level and high-level building blocks for Machine Learning / AI researchers and practitioners. It simplifies and streamlines work on deep learning models for healthcare and life sciences by offering tested components such as data loaders, pre-processing tools, deep learning models, and cloud integration utilities. The repository includes two Python packages, 'hi-ml-azure' for helper functions in AzureML, 'hi-ml' for ML components, and 'hi-ml-cpath' for models and workflows related to histopathology images.
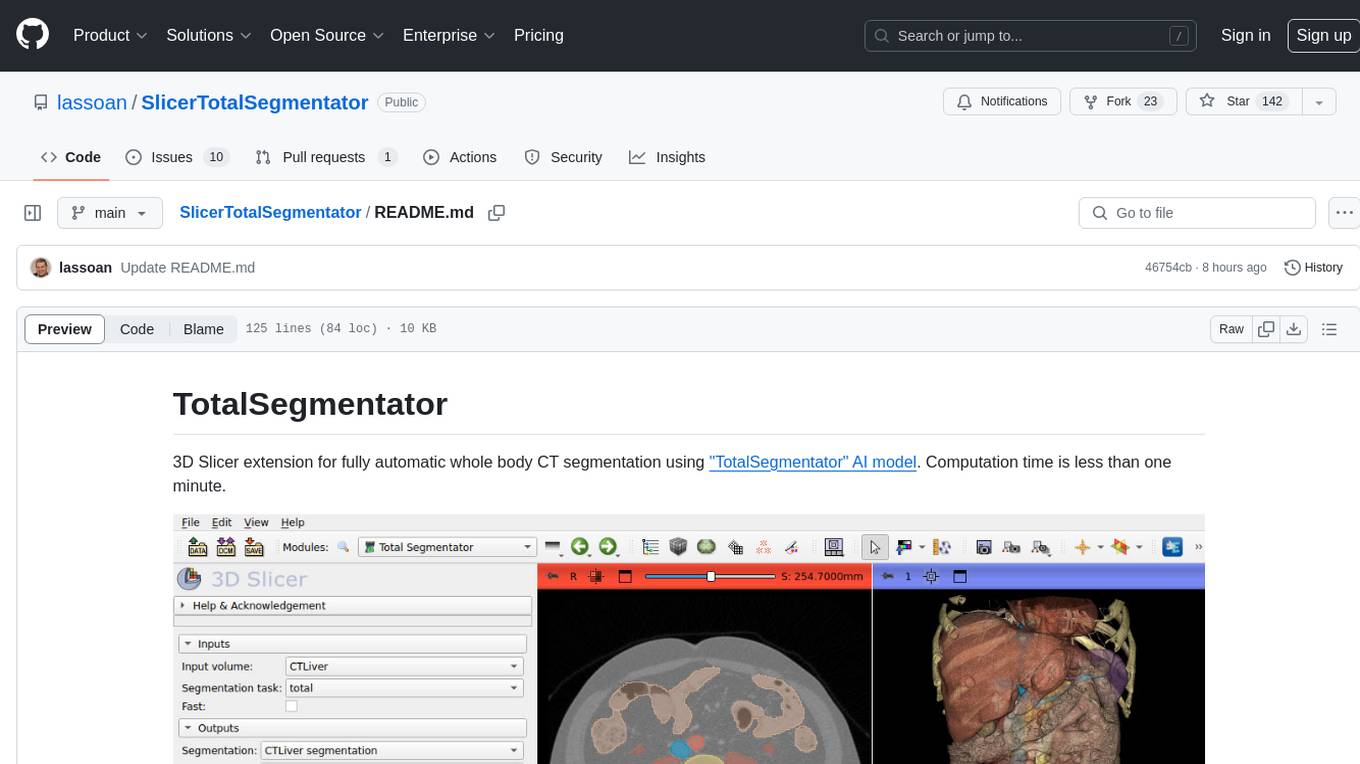
SlicerTotalSegmentator
TotalSegmentator is a 3D Slicer extension designed for fully automatic whole body CT segmentation using the 'TotalSegmentator' AI model. The computation time is less than one minute, making it efficient for research purposes. Users can set up GPU acceleration for faster segmentation. The tool provides a user-friendly interface for loading CT images, creating segmentations, and displaying results in 3D. Troubleshooting steps are available for common issues such as failed computation, GPU errors, and inaccurate segmentations. Contributions to the extension are welcome, following 3D Slicer contribution guidelines.
md-agent
MD-Agent is a LLM-agent based toolset for Molecular Dynamics. It uses Langchain and a collection of tools to set up and execute molecular dynamics simulations, particularly in OpenMM. The tool assists in environment setup, installation, and usage by providing detailed steps. It also requires API keys for certain functionalities, such as OpenAI and paper-qa for literature searches. Contributions to the project are welcome, with a detailed Contributor's Guide available for interested individuals.