
MOOSE
MOOSE (Multi-organ objective segmentation) a data-centric AI solution that generates multilabel organ segmentations to facilitate systemic TB whole-person research.The pipeline is based on nn-UNet and has the capability to segment 120 unique tissue classes from a whole-body 18F-FDG PET/CT image.
Stars: 182
MOOSE 2.0 is a leaner, meaner, and stronger tool for 3D medical image segmentation. It is built on the principles of data-centric AI and offers a wide range of segmentation models for both clinical and preclinical settings. MOOSE 2.0 is also versatile, allowing users to use it as a command-line tool for batch processing or as a library package for individual processing in Python projects. With its improved speed, accuracy, and flexibility, MOOSE 2.0 is the go-to tool for segmentation tasks.
README:
Welcome to the new and improved MOOSE (v3.0), where speed and efficiency aren't just buzzwords—they're a way of life.
💨 3x Faster Than Before
Like a moose sprinting through the woods (okay, maybe not that fast), MOOSE 3.0 is built for speed. It's 3x faster than its older sibling, MOOSE 2.0, which was already no slouch. Blink and you'll miss it. ⚡
💻 Memory: Light as a Feather, Strong as a Bull
Forget "Does it fit on my laptop?" The answer is YES. 🕺 Thanks to Dask wizardry, all that data stays in memory. No disk writes, no fuss. Run total-body CT on that 'decent' laptop you bought three years ago and feel like you’ve upgraded. 🥳
🛠️ Any OS, Anytime, Anywhere
Windows, Mac, Linux—we don’t play favorites. 🍏 Mac users, you’re in luck: MOOSE runs natively on MPS, getting you GPU-like speeds without the NVIDIA guilt. 🚀
🎯 Trained to Perfection
This is our best model yet, trained on a whopping 1.7k datasets. More data, better results. Plus you can run multiple models at the same time - You'll be slicing through images like a knife through warm butter. (Or tofu, if you prefer.) 🧈🔪
🖥️ The 'Herd' Mode 🖥️
Got a powerhouse server just sitting around? Time to let the herd loose! Flip the Herd Mode switch and watch MOOSE multiply across your compute like... well, like a herd of moose! 🦌🦌🦌 The more hardware you have, the faster your inference gets done. Scale up, speed up, and make every bit of your server earn its oats. 🌾💨
MOOSE 3.0 isn't just an upgrade—it's a lifestyle. A faster, leaner, and stronger lifestyle. Ready to join the herd? 🦌✨
MOOSE 3.0 offers a wide range of segmentation models catering to various clinical and preclinical needs. Here are the models currently available:
| Model Name | Intensities and Regions |
|---|---|
clin_ct_body |
1:Legs, 2:Body, 3:Head, 4:Arms |
clin_ct_cardiac |
1:heart_myocardium, 2:heart_atrium_left, 3:heart_ventricle_left, 4:heart_atrium_right, 5:heart_ventricle_right, 6:pulmonary_artery, 7:iliac_artery_left, 8:iliac_artery_right, 9:iliac_vena_left, 10:iliac_vena_right |
clin_ct_digestive |
1:esophagus, 2:trachea, 3:small_bowel, 4:duodenum, 5:colon, 6:urinary_bladder, 7:face |
clin_ct_fat |
1:spinal_chord, 2:skeletal_muscle, 3:subcutaneous_fat, 4:visceral_fat, 5:thoracic_fat, 6:eyes, 7:testicles, 8:prostate |
clin_ct_lungs |
1:lung_upper_lobe_left, 2:lung_lower_lobe_left, 3:lung_upper_lobe_right, 4:lung_middle_lobe_right, 5:lung_lower_lobe_right |
clin_ct_muscles |
1:gluteus_maximus_left, 2:gluteus_maximus_right, 3:gluteus_medius_left, 4:gluteus_medius_right, 5:gluteus_minimus_left, 6:gluteus_minimus_right, 7:autochthon_left, 8:autochthon_right, 9:iliopsoas_left, 10:iliopsoas_right |
clin_ct_organs |
1:spleen, 2:kidney_right, 3:kidney_left, 4:gallbladder, 5:liver, 6:stomach, 7:aorta, 8:inferior_vena_cava, 9:portal_vein_and_splenic_vein, 10:pancreas, 11:adrenal_gland_right, 12:adrenal_gland_left, 13:lung_upper_lobe_left, 14:lung_lower_lobe_left, 15:lung_upper_lobe_right, 16:lung_middle_lobe_right, 17:lung_lower_lobe_right |
clin_ct_peripheral_bones |
1:carpal_left, 2:carpal_right, 3:clavicle_left, 4:clavicle_right, 5:femur_left, 6:femur_right, 7:fibula_left, 8:fibula_right, 9:humerus_left, 10:humerus_right, 11:metacarpal_left, 12:metacarpal_right, 13:metatarsal_left, 14:metatarsal_right, 15:patella_left, 16:patella_right, 17:fingers_left, 18:fingers_right, 19:radius_left, 20:radius_right, 21:scapula_left, 22:scapula_right, 23:skull, 24:tarsal_left, 25:tarsal_right, 26:tibia_left, 27:tibia_right, 28:toes_left, 29:toes_right, 30:ulna_left, 31:ulna_right, 32:thyroid_left, 33:thyroid_right, 34:bladder |
clin_ct_ribs |
1:rib_left_1, 2:rib_left_2, 3:rib_left_3, 4:rib_left_4, 5:rib_left_5, 6:rib_left_6, 7:rib_left_7, 8:rib_left_8, 9:rib_left_9, 10:rib_left_10, 11:rib_left_11, 12:rib_left_12, 13:rib_right_1, 14:rib_right_2, 15:rib_right_3, 16:rib_right_4, 17:rib_right_5, 18:rib_right_6, 19:rib_right_7, 20:rib_right_8, 21:rib_right_9, 22:rib_right_10, 23:rib_right_11, 24:rib_right_12, 25:sternum |
clin_ct_vertebrae |
1:vertebra_C1, 2:vertebra_C2, 3:vertebra_C3, 4:vertebra_C4, 5:vertebra_C5, 6:vertebra_C6, 7:vertebra_C7, 8:vertebra_T1, 9:vertebra_T2, 10:vertebra_T3, 11:vertebra_T4, 12:vertebra_T5, 13:vertebra_T6, 14:vertebra_T7, 15:vertebra_T8, 16:vertebra_T9, 17:vertebra_T10, 18:vertebra_T11, 19:vertebra_T12, 20:vertebra_L1, 21:vertebra_L2, 22:vertebra_L3, 23:vertebra_L4, 24:vertebra_L5, 25:vertebra_S1, 26:hip_left, 27:hip_right, 28:sacrum |
| Model Name | Intensities and Regions |
|---|---|
preclin_ct_legs |
1:right_leg_muscle, 2:left_leg_muscle |
preclin_mr_all |
1:Brain, 2:Liver, 3:Intestines, 4:Pancreas, 5:Thyroid, 6:Spleen, 7:Bladder, 8:OuterKidney, 9:InnerKidney, 10:HeartInside, 11:HeartOutside, 12:WAT Subcutaneous, 13:WAT Visceral, 14:BAT, 15:Muscle TF, 16:Muscle TB, 17:Muscle BB, 18:Muscle BF, 19:Aorta, 20:Lung, 21:Stomach |
Each model is designed to provide high-quality segmentation with MOOSE 3.0's optimized algorithms and data-centric AI principles.
- Shiyam Sundar, L. K., Yu, J., Muzik, O., Kulterer, O., Fueger, B. J., Kifjak, D., Nakuz, T., Shin, H. M., Sima, A. K., Kitzmantl, D., Badawi, R. D., Nardo, L., Cherry, S. R., Spencer, B. A., Hacker, M., & Beyer, T. (2022). Fully-automated, semantic segmentation of whole-body 18F-FDG PET/CT images based on data-centric artificial intelligence. Journal of Nuclear Medicine. https://doi.org/10.2967/jnumed.122.264063
- Isensee, F., Jaeger, P.F., Kohl, S.A.A. et al. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 18, 203–211 (2021). https://doi.org/10.1038/s41592-020-01008-z
Before you dive into the incredible world of MOOSE 3.0, here are a few things you need to ensure for an optimal experience:
-
Operating System: We've got you covered whether you're on Windows, Mac, or Linux. MOOSE 3.0 has been tested across these platforms to ensure seamless operation.
-
Memory: MOOSE 3.0 has quite an appetite! Make sure you have at least 16GB of RAM for the smooth running of all tasks.
-
GPU: If speed is your game, an NVIDIA GPU is the name! MOOSE 3.0 leverages GPU acceleration to deliver results fast. Don't worry if you don't have one, though - it will still work, just at a slower pace.
-
Python: Ensure that you have Python 3.10 installed on your system. MOOSE 3.0 likes to keep up with the latest, after all!
So, that's it! Make sure you're geared up with these specifications, and you're all set to explore everything MOOSE 3.0 has to offer. 🚀🌐
Available on Windows, Linux, and MacOS, the installation is as simple as it gets. Follow our step-by-step guide below and set sail on your journey with MOOSE 3.0.
-
First, create a Python environment. You can name it to your liking; for example, 'moose-env'.
python3.10 -m venv moose-env
-
Activate your newly created environment.
source moose-env/bin/activate # for Linux
-
Install MOOSE 3.0.
pip install moosez
Voila! You're all set to explore with MOOSE 3.0.
-
First, create a Python environment. You can name it to your liking; for example, 'moose-env'.
python3.10 -m venv moose-env
-
Activate your newly created environment.
source moose-env/bin/activate -
Install MOOSE 3.0 and a special fork of PyTorch (MPS specific). You need to install the MPS specific branch for making MOOSE work with MPS
pip install moosez pip install git+https://github.com/LalithShiyam/pytorch-mps.git
Now you are ready to use MOOSE on Apple Silicon 🏎⚡️.
-
Create a Python environment. You could name it 'moose-env', or as you wish.
python3.10 -m venv moose-env
-
Activate your newly created environment.
.\moose-env\Scripts\activate
-
Go to the PyTorch website and install the appropriate PyTorch version for your system. !DO NOT SKIP THIS!
-
Finally, install MOOSE 3.0.
pip install moosez
There you have it! You're ready to venture into the world of 3D medical image segmentation with MOOSE 3.0.
Happy exploring! 🚀🔬
Getting started with MOOSE 3.0 is as easy as slicing through butter 🧈🔪. Use the command-line tool to process multiple segmentation models in sequence or in parallel, making your workflow a breeze. 🌬️
You can now run single or several models in sequence with a single command. Just provide the path to your subject images and list the segmentation models you wish to apply:
# For single model inference
moosez -d <path_to_image_dir> -m <model_name>
# For multiple model inference
moosez -d <path_to_image_dir> \
-m <model_name1> \
<model_name2> \
<model_name3> \For instance, to run clinical CT organ segmentation on a directory of images, you can use the following command:
moosez -d <path_to_image_dir> -m clin_ct_organsLikewise, to run multiple models e.g. organs, ribs, and vertebrae, you can use the following command:
moosez -d <path_to_image_dir> \
-m clin_ct_organs \
clin_ct_ribs \
clin_ct_vertebraeMOOSE 3.0 will handle each model one after the other—no fuss, no hassle. 🙌✨
Got a powerful server or HPC? Let the herd roam! 🦌🚀 Use Herd Mode to run multiple MOOSE instances in parallel. Just add the -herd flag with the number of instances you wish to run simultaneously:
moosez -d <path_to_image_dir> \
-m clin_ct_organs \
clin_ct_ribs \
clin_ct_vertebrae \
-herd 2MOOSE will run two instances at the same time, utilizing your compute power like a true multitasking pro. 💪👨💻👩💻
And that's it! MOOSE 3.0 lets you process with ease and speed. ⚡✨
Need assistance along the way? Don't worry, we've got you covered. Simply type:
moosez -hThis command will provide you with all the help and the information about the available models and the regions it segments.
MOOSE 3.0 isn't just a command-line powerhouse; it’s also a flexible library for Python projects. Here’s how to make the most of it:
First, import the moose function from the moosez package in your Python script:
from moosez import mooseThe moose function is versatile and accepts various input types. It takes four main arguments:
-
input: The data to process, which can be:- A path to an input file or directory (NIfTI, either
.niior.nii.gz). - A tuple containing a NumPy array and its spacing (e.g.,
numpy_array,(spacing_x, spacing_y, spacing_z)). - A
SimpleITKimage object.
- A path to an input file or directory (NIfTI, either
-
model_names: A single model name or a list of model names for segmentation. -
output_dir: The directory where the results will be saved. -
accelerator: The type of accelerator to use ("cpu","cuda", or"mps"for Mac).
Here are some examples to illustrate different ways to use the moose function:
-
Using a file path and multiple models:
moose('/path/to/input/file', ['clin_ct_organs', 'clin_ct_ribs'], '/path/to/save/output', 'cuda')
-
Using a NumPy array with spacing:
moose((numpy_array, (1.5, 1.5, 1.5)), 'clin_ct_organs', '/path/to/save/output', 'cuda')
-
Using a SimpleITK image:
moose(simple_itk_image, 'clin_ct_organs', '/path/to/save/output', 'cuda')
That's it! With these flexible inputs, you can use MOOSE 3.0 to fit your workflow perfectly—whether you’re processing a single image, a stack of files, or leveraging different data formats. 🖥️🎉
Happy segmenting with MOOSE 3.0! 🦌💫
Using MOOSE 3.0 optimally requires your data to be structured according to specific conventions. MOOSE 3.0 supports both DICOM and NIFTI formats. For DICOM files, MOOSE infers the modality from the DICOM tags and checks if the given modality is suitable for the chosen segmentation model. However, for NIFTI files, users need to ensure that the files are named with the correct modality as a suffix.
Please structure your dataset as follows:
MOOSEv2_data/ 📁
├── S1 📂
│ ├── AC-CT 📂
│ │ ├── WBACCTiDose2_2001_CT001.dcm 📄
│ │ ├── WBACCTiDose2_2001_CT002.dcm 📄
│ │ ├── ... 🗂️
│ │ └── WBACCTiDose2_2001_CT532.dcm 📄
│ └── AC-PT 📂
│ ├── DetailWB_CTACWBPT001_PT001.dcm 📄
│ ├── DetailWB_CTACWBPT001_PT002.dcm 📄
│ ├── ... 🗂️
│ └── DetailWB_CTACWBPT001_PT532.dcm 📄
├── S2 📂
│ └── CT_S2.nii 📄
├── S3 📂
│ └── CT_S3.nii 📄
├── S4 📂
│ └── S4_ULD_FDG_60m_Dynamic_Patlak_HeadNeckThoAbd_20211025075852_2.nii 📄
└── S5 📂
└── CT_S5.nii 📄
Note: If the necessary naming conventions are not followed, MOOSE 3.0 will skip the subjects.
When using NIFTI files, you should name the file with the appropriate modality as a suffix.
For instance, if you have chosen the model_name as clin_ct_organs, the CT scan for subject 'S2' in NIFTI format, should have the modality tag 'CT_' attached to the file name, e.g. CT_S2.nii. In the directory shown above, every subject will be processed by moosez except S4.
Remember: Adhering to these file naming and directory structure conventions ensures smooth and efficient processing with MOOSE 3.0. Happy segmenting! 🚀
Want to power-up your medical image segmentation tasks? ⚡ Join the MooseZ community and contribute your own nnUNetv2 models! 🥇:
By adding your custom models to MooseZ, you can enjoy:
- ⏩ Increased Speed - MooseZ is optimized for fast performance. Use it to get your results faster!
- 💾 Reduced Memory - MooseZ is designed to be efficient and lean, so it uses less memory!
So why wait? Make your models fly with MooseZ
-
Prepare Your Model 📁
Train your model using
nnUNetv2and get it ready for the big leagues! -
Update AVAILABLE_MODELS List ✏️
Include your model's unique identifier to the
AVAILABLE_MODELSlist in the resources.py file. The model name should follow a specific syntax: 'clin' or 'preclin' (indicating Clinical or Preclinical), modality tag (like 'ct', 'pt', 'mr'), and then the tissue of interest. -
Update MODELS Dictionary 📋
Add a new entry to the
MODELSdictionary in the resources.py file. Fill in the corresponding details (like URL, filename, directory, trainer type, voxel spacing, and multilabel prefix). -
Update expected_modality Function 📝
Update the
expected_modalityfunction in the resources.py file to return the imaging technique, modality, and tissue of interest for your model. -
Update map_model_name_to_task_number Function 🗺️
Modify the
map_model_name_to_task_numberfunction in the resources.py file to return the task number associated with your model. -
Update
ORGAN_INDICESinconstants.py🧠Append the
ORGAN_INDICESdictionary in the constants.py with your label intensity to region mapping. This is particularly important if you would like to have your stats from the PET images based on your CT masks.
That's it! You've successfully contributed your own model to the MooseZ community! 🎊
With your contribution 🙋, MooseZ becomes a stronger and more robust tool for medical image segmentation! 💪
All of our Python packages here at QIMP carry a special signature – a distinctive 'Z' at the end of their names. The 'Z' is more than just a letter to us; it's a symbol of our forward-thinking approach and commitment to continuous innovation.
Our MOOSE package, for example, is named as 'moosez', pronounced "moose-see". So, why 'Z'?
Well, in the world of mathematics and science, 'Z' often represents the unknown, the variable that's yet to be discovered, or the final destination in a series. We at QIMP believe in always pushing boundaries, venturing into uncharted territories, and staying on the cutting edge of technology. The 'Z' embodies this philosophy. It represents our constant quest to uncover what lies beyond the known, to explore the undiscovered, and to bring you the future of medical imaging.
Each time you see a 'Z' in one of our package names, be reminded of the spirit of exploration and discovery that drives our work. With QIMP, you're not just installing a package; you're joining us on a journey to the frontiers of medical image processing. Here's to exploring the 'Z' dimension together! 🚀
🦌 MOOSE: A part of the enhance.pet community
</tr>
 Lalith Kumar Shiyam Sundar 💻 📖 |
 Sebastian Gutschmayer 💻 |
n7k-dobri 💻 |
Manuel Pires 💻 |
Zach Chalampalakis 💻 |
David Haberl 💻 |
W7ebere 📖 |
|
Kazezaka 💻 |
Loic Tetrel @ Kitware 💻 📖 |
For Tasks:
Click tags to check more tools for each tasksFor Jobs:
Alternative AI tools for MOOSE
Similar Open Source Tools

MOOSE
MOOSE 2.0 is a leaner, meaner, and stronger tool for 3D medical image segmentation. It is built on the principles of data-centric AI and offers a wide range of segmentation models for both clinical and preclinical settings. MOOSE 2.0 is also versatile, allowing users to use it as a command-line tool for batch processing or as a library package for individual processing in Python projects. With its improved speed, accuracy, and flexibility, MOOSE 2.0 is the go-to tool for segmentation tasks.
LLM-Pruner
LLM-Pruner is a tool for structural pruning of large language models, allowing task-agnostic compression while retaining multi-task solving ability. It supports automatic structural pruning of various LLMs with minimal human effort. The tool is efficient, requiring only 3 minutes for pruning and 3 hours for post-training. Supported LLMs include Llama-3.1, Llama-3, Llama-2, LLaMA, BLOOM, Vicuna, and Baichuan. Updates include support for new LLMs like GQA and BLOOM, as well as fine-tuning results achieving high accuracy. The tool provides step-by-step instructions for pruning, post-training, and evaluation, along with a Gradio interface for text generation. Limitations include issues with generating repetitive or nonsensical tokens in compressed models and manual operations for certain models.
quivr
Quivr is a personal assistant powered by Generative AI, designed to be a second brain for users. It offers fast and efficient access to data, ensuring security and compatibility with various file formats. Quivr is open source and free to use, allowing users to share their brains publicly or keep them private. The marketplace feature enables users to share and utilize brains created by others, boosting productivity. Quivr's offline mode provides anytime, anywhere access to data. Key features include speed, security, OS compatibility, file compatibility, open source nature, public/private sharing options, a marketplace, and offline mode.
allms
allms is a versatile and powerful library designed to streamline the process of querying Large Language Models (LLMs). Developed by Allegro engineers, it simplifies working with LLM applications by providing a user-friendly interface, asynchronous querying, automatic retrying mechanism, error handling, and output parsing. It supports various LLM families hosted on different platforms like OpenAI, Google, Azure, and GCP. The library offers features for configuring endpoint credentials, batch querying with symbolic variables, and forcing structured output format. It also provides documentation, quickstart guides, and instructions for local development, testing, updating documentation, and making new releases.

MInference
MInference is a tool designed to accelerate pre-filling for long-context Language Models (LLMs) by leveraging dynamic sparse attention. It achieves up to a 10x speedup for pre-filling on an A100 while maintaining accuracy. The tool supports various decoding LLMs, including LLaMA-style models and Phi models, and provides custom kernels for attention computation. MInference is useful for researchers and developers working with large-scale language models who aim to improve efficiency without compromising accuracy.
LightAgent
LightAgent is a lightweight, open-source Agentic AI development framework with memory, tools, and a tree of thought. It supports multi-agent collaboration, autonomous learning, tool integration, complex task handling, and multi-model support. It also features a streaming API, tool generator, agent self-learning, adaptive tool mechanism, and more. LightAgent is designed for intelligent customer service, data analysis, automated tools, and educational assistance.
LightAgent
LightAgent is a lightweight, open-source active Agentic AI development framework with memory, tools, and a tree of thought. It supports multi-agent collaboration, autonomous learning, tool integration, complex goals, and multi-model support. It enables simpler self-learning agents, seamless integration with major chat frameworks, and quick tool generation. LightAgent also supports memory modules, tool integration, tree of thought planning, multi-agent collaboration, streaming API, agent self-learning, Langfuse log tracking, and agent assessment. It is compatible with various large models and offers features like intelligent customer service, data analysis, automated tools, and educational assistance.
OpenMusic
OpenMusic is a repository providing an implementation of QA-MDT, a Quality-Aware Masked Diffusion Transformer for music generation. The code integrates state-of-the-art models and offers training strategies for music generation. The repository includes implementations of AudioLDM, PixArt-alpha, MDT, AudioMAE, and Open-Sora. Users can train or fine-tune the model using different strategies and datasets. The model is well-pretrained and can be used for music generation tasks. The repository also includes instructions for preparing datasets, training the model, and performing inference. Contact information is provided for any questions or suggestions regarding the project.
Easy-Translate
Easy-Translate is a script designed for translating large text files with a single command. It supports various models like M2M100, NLLB200, SeamlessM4T, LLaMA, and Bloom. The tool is beginner-friendly and offers seamless and customizable features for advanced users. It allows acceleration on CPU, multi-CPU, GPU, multi-GPU, and TPU, with support for different precisions and decoding strategies. Easy-Translate also provides an evaluation script for translations. Built on HuggingFace's Transformers and Accelerate library, it supports prompt usage and loading huge models efficiently.

AirGym
AirGym is an open source Python quadrotor simulator based on IsaacGym, providing a high-fidelity dynamics and Deep Reinforcement Learning (DRL) framework for quadrotor robot learning research. It offers a lightweight and customizable platform with strict alignment with PX4 logic, multiple control modes, and Sim-to-Real toolkits. Users can perform tasks such as Hovering, Balloon, Tracking, Avoid, and Planning, with the ability to create customized environments and tasks. The tool also supports training from scratch, visual encoding approaches, playing and testing of trained models, and customization of new tasks and assets.
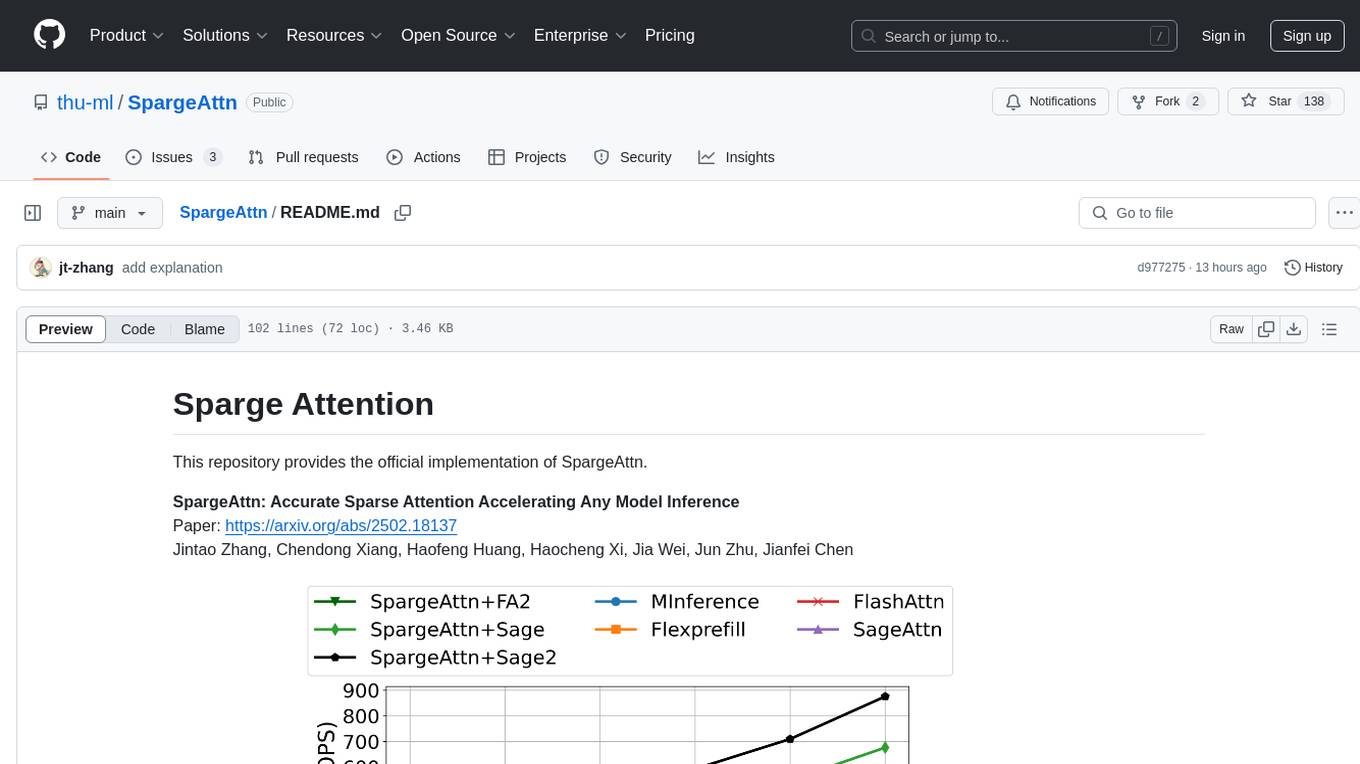
SpargeAttn
SpargeAttn is an official implementation designed for accelerating any model inference by providing accurate sparse attention. It offers a significant speedup in model performance while maintaining quality. The tool is based on SageAttention and SageAttention2, providing options for different levels of optimization. Users can easily install the package and utilize the available APIs for their specific needs. SpargeAttn is particularly useful for tasks requiring efficient attention mechanisms in deep learning models.
qa-mdt
This repository provides an implementation of QA-MDT, integrating state-of-the-art models for music generation. It offers a Quality-Aware Masked Diffusion Transformer for enhanced music generation. The code is based on various repositories like AudioLDM, PixArt-alpha, MDT, AudioMAE, and Open-Sora. The implementation allows for training and fine-tuning the model with different strategies and datasets. The repository also includes instructions for preparing datasets in LMDB format and provides a script for creating a toy LMDB dataset. The model can be used for music generation tasks, with a focus on quality injection to enhance the musicality of generated music.
pandas-ai
PandasAI is a Python library that makes it easy to ask questions to your data in natural language. It helps you to explore, clean, and analyze your data using generative AI.
chromem-go
chromem-go is an embeddable vector database for Go with a Chroma-like interface and zero third-party dependencies. It enables retrieval augmented generation (RAG) and similar embeddings-based features in Go apps without the need for a separate database. The focus is on simplicity and performance for common use cases, allowing querying of documents with minimal memory allocations. The project is in beta and may introduce breaking changes before v1.0.0.
raid
RAID is the largest and most comprehensive dataset for evaluating AI-generated text detectors. It contains over 10 million documents spanning 11 LLMs, 11 genres, 4 decoding strategies, and 12 adversarial attacks. RAID is designed to be the go-to location for trustworthy third-party evaluation of popular detectors. The dataset covers diverse models, domains, sampling strategies, and attacks, making it a valuable resource for training detectors, evaluating generalization, protecting against adversaries, and comparing to state-of-the-art models from academia and industry.

embodied-agents
Embodied Agents is a toolkit for integrating large multi-modal models into existing robot stacks with just a few lines of code. It provides consistency, reliability, scalability, and is configurable to any observation and action space. The toolkit is designed to reduce complexities involved in setting up inference endpoints, converting between different model formats, and collecting/storing datasets. It aims to facilitate data collection and sharing among roboticists by providing Python-first abstractions that are modular, extensible, and applicable to a wide range of tasks. The toolkit supports asynchronous and remote thread-safe agent execution for maximal responsiveness and scalability, and is compatible with various APIs like HuggingFace Spaces, Datasets, Gymnasium Spaces, Ollama, and OpenAI. It also offers automatic dataset recording and optional uploads to the HuggingFace hub.
For similar tasks

MOOSE
MOOSE 2.0 is a leaner, meaner, and stronger tool for 3D medical image segmentation. It is built on the principles of data-centric AI and offers a wide range of segmentation models for both clinical and preclinical settings. MOOSE 2.0 is also versatile, allowing users to use it as a command-line tool for batch processing or as a library package for individual processing in Python projects. With its improved speed, accuracy, and flexibility, MOOSE 2.0 is the go-to tool for segmentation tasks.
For similar jobs

MOOSE
MOOSE 2.0 is a leaner, meaner, and stronger tool for 3D medical image segmentation. It is built on the principles of data-centric AI and offers a wide range of segmentation models for both clinical and preclinical settings. MOOSE 2.0 is also versatile, allowing users to use it as a command-line tool for batch processing or as a library package for individual processing in Python projects. With its improved speed, accuracy, and flexibility, MOOSE 2.0 is the go-to tool for segmentation tasks.
nitrain
Nitrain is a framework for medical imaging AI that provides tools for sampling and augmenting medical images, training models on medical imaging datasets, and visualizing model results in a medical imaging context. It supports using pytorch, keras, and tensorflow.





&color=9400D3&style=flat-square&logo=python)
&color=9400D3&style=flat-square&logo=python)


