
facet
Human-explainable AI.
Stars: 525
FACET is an open source library for human-explainable AI that combines model inspection and model-based simulation to provide better explanations for supervised machine learning models. It offers an efficient and transparent machine learning workflow, enhancing scikit-learn's pipelining paradigm with new capabilities for model selection, inspection, and simulation. FACET introduces new algorithms for quantifying dependencies and interactions between features in ML models, as well as for conducting virtual experiments to optimize predicted outcomes. The tool ensures end-to-end traceability of features using an augmented version of scikit-learn with enhanced support for pandas data frames. FACET also provides model inspection methods for scikit-learn estimators, enhancing global metrics like synergy and redundancy to complement the local perspective of SHAP. Additionally, FACET offers model simulation capabilities for conducting univariate uplift simulations based on important features like BMI.
README:
.. image:: sphinx/source/_images/Gamma_Facet_Logo_RGB_LB.svg
FACET is an open source library for human-explainable AI. It combines sophisticated model inspection and model-based simulation to enable better explanations of your supervised machine learning models.
FACET is composed of the following key components:
+-----------------+-----------------------------------------------------------------------+
| |spacer| | Model Inspection |
| | |
| |inspect| | FACET introduces a new algorithm to quantify dependencies and |
| | interactions between features in ML models. |
| | This new tool for human-explainable AI adds a new, global |
| | perspective to the observation-level explanations provided by the |
| | popular SHAP <https://shap.readthedocs.io/en/stable/>__ approach. |
| | To learn more about FACET’s model inspection capabilities, see the |
| | getting started example below. |
+-----------------+-----------------------------------------------------------------------+
| |spacer| | Model Simulation |
| | |
| |sim| | FACET’s model simulation algorithms use ML models for |
| | virtual experiments to help identify scenarios that optimise |
| | predicted outcomes. |
| | To quantify the uncertainty in simulations, FACET utilises a range |
| | of bootstrapping algorithms including stationary and stratified |
| | bootstraps. |
| | For an example of FACET’s bootstrap simulations, see the |
| | quickstart example below. |
+-----------------+-----------------------------------------------------------------------+
| |spacer| | Enhanced Machine Learning Workflow |
| | |
| |pipe| | FACET offers an efficient and transparent machine learning |
| | workflow, enhancing |
| | scikit-learn <https://scikit-learn.org/stable/index.html>'s |
| | tried and tested pipelining paradigm with new capabilities for model |
| | selection, inspection, and simulation. |
| | FACET also introduces |
| | sklearndf <https://github.com/BCG-X-Official/sklearndf> |
| | [documentation <https://bcg-x-official.github.io/sklearndf/index.html>__]|
| | an augmented version of scikit-learn with enhanced support for |
| | pandas data frames that ensures end-to-end traceability of features.|
+-----------------+-----------------------------------------------------------------------+
.. Begin-Badges
|pypi| |conda| |azure_build| |azure_code_cov| |python_versions| |code_style| |made_with_sphinx_doc| |License_badge|
.. End-Badges
FACET supports both PyPI and Anaconda. We recommend to install FACET into a dedicated environment.
Anaconda
.. code-block:: sh
conda create -n facet
conda activate facet
conda install -c bcg_gamma -c conda-forge gamma-facet
Pip
~~~
macOS and Linux:
^^^^^^^^^^^^^^^^
.. code-block:: sh
python -m venv facet
source facet/bin/activate
pip install gamma-facet
Windows:
^^^^^^^^
.. code-block:: dosbatch
python -m venv facet
facet\Scripts\activate.bat
pip install gamma-facet
Quickstart
----------
The following quickstart guide provides a minimal example workflow to get you
up and running with FACET.
For additional tutorials and the API reference,
see the `FACET documentation <https://bcg-x-official.github.io/facet/docs-version/2-0>`__.
Changes and additions to new versions are summarized in the
`release notes <https://bcg-x-official.github.io/facet/docs-version/2-0/release_notes.html>`__.
Enhanced Machine Learning Workflow
To demonstrate the model inspection capability of FACET, we first create a
pipeline to fit a learner. In this simple example we will use the
diabetes dataset <https://web.stanford.edu/~hastie/Papers/LARS/diabetes.data>__
which contains age, sex, BMI and blood pressure along with 6 blood serum
measurements as features. This dataset was used in this
publication <https://statweb.stanford.edu/~tibs/ftp/lars.pdf>.
A transformed version of this dataset is also available on scikit-learn
here <https://scikit-learn.org/stable/datasets/toy_dataset.html#diabetes-dataset>.
In this quickstart we will train a Random Forest regressor using 10 repeated
5-fold CV to predict disease progression after one year. With the use of
sklearndf we can create a pandas DataFrame compatible workflow. However,
FACET provides additional enhancements to keep track of our feature matrix
and target vector using a sample object (Sample) and easily compare
hyperparameter configurations and even multiple learners with the LearnerSelector.
.. code-block:: Python
# standard imports
import pandas as pd
from sklearn.model_selection import RepeatedKFold, GridSearchCV
# some helpful imports from sklearndf
from sklearndf.pipeline import RegressorPipelineDF
from sklearndf.regression import RandomForestRegressorDF
# relevant FACET imports
from facet.data import Sample
from facet.selection import LearnerSelector, ParameterSpace
# declaring url with data
data_url = 'https://web.stanford.edu/~hastie/Papers/LARS/diabetes.data'
#importing data from url
diabetes_df = pd.read_csv(data_url, delimiter='\t').rename(
# renaming columns for better readability
columns={
'S1': 'TC', # total serum cholesterol
'S2': 'LDL', # low-density lipoproteins
'S3': 'HDL', # high-density lipoproteins
'S4': 'TCH', # total cholesterol/ HDL
'S5': 'LTG', # lamotrigine level
'S6': 'GLU', # blood sugar level
'Y': 'Disease_progression' # measure of progress since 1yr of baseline
}
)
# create FACET sample object
diabetes_sample = Sample(observations=diabetes_df, target_name="Disease_progression")
# create a (trivial) pipeline for a random forest regressor
rnd_forest_reg = RegressorPipelineDF(
regressor=RandomForestRegressorDF(n_estimators=200, random_state=42)
)
# define parameter space for models which are "competing" against each other
rnd_forest_ps = ParameterSpace(rnd_forest_reg)
rnd_forest_ps.regressor.min_samples_leaf = [8, 11, 15]
rnd_forest_ps.regressor.max_depth = [4, 5, 6]
# create repeated k-fold CV iterator
rkf_cv = RepeatedKFold(n_splits=5, n_repeats=10, random_state=42)
# rank your candidate models by performance
selector = LearnerSelector(
searcher_type=GridSearchCV,
parameter_space=rnd_forest_ps,
cv=rkf_cv,
n_jobs=-3,
scoring="r2"
).fit(diabetes_sample)
# get summary report
selector.summary_report()
.. image:: sphinx/source/_images/ranker_summary.png :width: 600
We can see based on this minimal workflow that a value of 11 for minimum samples in the leaf and 5 for maximum tree depth was the best performing of the three considered values. This approach easily extends to additional hyperparameters for the learner, and for multiple learners.
Model Inspection
FACET implements several model inspection methods for
`scikit-learn <https://scikit-learn.org/stable/index.html>`__ estimators.
FACET enhances model inspection by providing global metrics that complement
the local perspective of SHAP (see
`[arXiv:2107.12436] <https://arxiv.org/abs/2107.12436>`__ for a formal description).
The key global metrics for each pair of features in a model are:
- **Synergy**
The degree to which the model combines information from one feature with
another to predict the target. For example, let's assume we are predicting
cardiovascular health using age and gender and the fitted model includes
a complex interaction between them. This means these two features are
synergistic for predicting cardiovascular health. Further, both features
are important to the model and removing either one would significantly
impact performance. Let's assume age brings more information to the joint
contribution than gender. This asymmetric contribution means the synergy for
(age, gender) is less than the synergy for (gender, age). To think about it another
way, imagine the prediction is a coordinate you are trying to reach.
From your starting point, age gets you much closer to this point than
gender, however, you need both to get there. Synergy reflects the fact
that gender gets more help from age (higher synergy from the perspective
of gender) than age does from gender (lower synergy from the perspective of
age) to reach the prediction. *This leads to an important point: synergy
is a naturally asymmetric property of the global information two interacting
features contribute to the model predictions.* Synergy is expressed as a
percentage ranging from 0% (full autonomy) to 100% (full synergy).
- **Redundancy**
The degree to which a feature in a model duplicates the information of a
second feature to predict the target. For example, let's assume we had
house size and number of bedrooms for predicting house price. These
features capture similar information as the more bedrooms the larger
the house and likely a higher price on average. The redundancy for
(number of bedrooms, house size) will be greater than the redundancy
for (house size, number of bedrooms). This is because house size
"knows" more of what number of bedrooms does for predicting house price
than vice-versa. Hence, there is greater redundancy from the perspective
of number of bedrooms. Another way to think about it is removing house
size will be more detrimental to model performance than removing number
of bedrooms, as house size can better compensate for the absence of
number of bedrooms. This also implies that house size would be a more
important feature than number of bedrooms in the model. *The important
point here is that like synergy, redundancy is a naturally asymmetric
property of the global information feature pairs have for predicting
an outcome.* Redundancy is expressed as a percentage ranging from 0%
(full uniqueness) to 100% (full redundancy).
.. code-block:: Python
# fit the model inspector
from facet.inspection import LearnerInspector
inspector = LearnerInspector(
pipeline=selector.best_estimator_,
n_jobs=-3
).fit(diabetes_sample)
**Synergy**
.. code-block:: Python
# visualise synergy as a matrix
from pytools.viz.matrix import MatrixDrawer
synergy_matrix = inspector.feature_synergy_matrix()
MatrixDrawer(style="matplot%").draw(synergy_matrix, title="Synergy Matrix")
.. image:: sphinx/source/_images/synergy_matrix.png
:width: 600
For any feature pair (A, B), the first feature (A) is the row, and the second
feature (B) the column. For example, looking across the row for `LTG` (Lamotrigine)
there is hardly any synergy with other features in the model (≤ 1%).
However, looking down the column for `LTG` (i.e., from the perspective of other features
relative with `LTG`) we find that many features (the rows) are aided by synergy with
with `LTG` (up to 27% in the case of LDL). We conclude that:
- `LTG` is a strongly autonomous feature, displaying minimal synergy with other
features for predicting disease progression after one year.
- The contribution of other features to predicting disease progression after one
year is partly enabled by the presence of `LTG`.
High synergy between pairs of features must be considered carefully when investigating
impact, as the values of both features jointly determine the outcome. It would not make
much sense to consider `LDL` without the context provided by `LTG` given close
to 27% synergy of `LDL` with `LTG` for predicting progression after one year.
**Redundancy**
.. code-block:: Python
# visualise redundancy as a matrix
redundancy_matrix = inspector.feature_redundancy_matrix()
MatrixDrawer(style="matplot%").draw(redundancy_matrix, title="Redundancy Matrix")
.. image:: sphinx/source/_images/redundancy_matrix.png
:width: 600
For any feature pair (A, B), the first feature (A) is the row, and the second feature
(B) the column. For example, if we look at the feature pair (`LDL`, `TC`) from the
perspective of `LDL` (Low-Density Lipoproteins), then we look-up the row for `LDL`
and the column for `TC` and find 38% redundancy. This means that 38% of the information
in `LDL` to predict disease progression is duplicated in `TC`. This
redundancy is the same when looking "from the perspective" of `TC` for (`TC`, `LDL`),
but need not be symmetrical in all cases (see `LTG` vs. `TCH`).
If we look at `TCH`, it has between 22–32% redundancy each with `LTG` and `HDL`, but
the same does not hold between `LTG` and `HDL` – meaning `TCH` shares different
information with each of the two features.
**Clustering redundancy**
As detailed above redundancy and synergy for a feature pair is from the
"perspective" of one of the features in the pair, and so yields two distinct
values. However, a symmetric version can also be computed that provides not
only a simplified perspective but allows the use of (1 - metric) as a
feature distance. With this distance hierarchical, single linkage clustering
is applied to create a dendrogram visualization. This helps to identify
groups of low distance, features which activate "in tandem" to predict the
outcome. Such information can then be used to either reduce clusters of
highly redundant features to a subset or highlight clusters of highly
synergistic features that should always be considered together.
Let's look at the example for redundancy.
.. code-block:: Python
# visualise redundancy using a dendrogram
from pytools.viz.dendrogram import DendrogramDrawer
redundancy = inspector.feature_redundancy_linkage()
DendrogramDrawer().draw(data=redundancy, title="Redundancy Dendrogram")
.. image:: sphinx/source/_images/redundancy_dendrogram.png
:width: 600
Based on the dendrogram we can see that the feature pairs (`LDL`, `TC`)
and (`HDL`, `TCH`) each represent a cluster in the dendrogram and that `LTG` and `BMI`
have the highest importance. As potential next actions we could explore the impact of
removing `TCH`, and one of `TC` or `LDL` to further simplify the model and obtain a
reduced set of independent features.
Please see the
`API reference <https://bcg-x-official.github.io/facet/apidoc/facet.html>`__
for more detail.
Model Simulation
Taking the BMI feature as an example of an important and highly independent feature,
we do the following for the simulation:
- We use FACET's
ContinuousRangePartitionerto split the range of observed values ofBMIinto intervals of equal size. Each partition is represented by the central value of that partition. - For each partition, the simulator creates an artificial copy of the original sample assuming the variable to be simulated has the same value across all observations – which is the value representing the partition. Using the best estimator acquired from the selector, the simulator now re-predicts all targets using the models trained for full sample and determines the uplift of the target variable resulting from this.
- The FACET
SimulationDrawerallows us to visualise the result; both in a matplotlib and a plain-text style.
.. code-block:: Python
# FACET imports
from facet.validation import BootstrapCV
from facet.simulation import UnivariateUpliftSimulator
from facet.data.partition import ContinuousRangePartitioner
from facet.simulation.viz import SimulationDrawer
# create bootstrap CV iterator
bscv = BootstrapCV(n_splits=1000, random_state=42)
SIM_FEAT = "BMI"
simulator = UnivariateUpliftSimulator(
model=selector.best_estimator_,
sample=diabetes_sample,
n_jobs=-3
)
# split the simulation range into equal sized partitions
partitioner = ContinuousRangePartitioner()
# run the simulation
simulation = simulator.simulate_feature(feature_name=SIM_FEAT, partitioner=partitioner)
# visualise results
SimulationDrawer().draw(data=simulation, title=SIM_FEAT)
.. image:: sphinx/source/_images/simulation_output.png
We would conclude from the figure that higher values of BMI are associated with
an increase in disease progression after one year, and that for a BMI of 28
and above, there is a significant increase in disease progression after one year
of at least 26 points.
FACET is stable and is being supported long-term.
Contributions to FACET are welcome and appreciated.
For any bug reports or feature requests/enhancements please use the appropriate
GitHub form <https://github.com/BCG-X-Official/facet/issues>_, and if you wish to do so,
please open a PR addressing the issue.
We do ask that for any major changes please discuss these with us first via an issue or using our team email: [email protected].
For further information on contributing please see our
contribution guide <https://bcg-x-official.github.io/facet/contribution_guide.html>__.
FACET is licensed under Apache 2.0 as described in the
LICENSE <https://github.com/BCG-X-Official/facet/blob/develop/LICENSE>_ file.
FACET is built on top of two popular packages for Machine Learning:
- The
scikit-learn <https://scikit-learn.org/stable/index.html>__ learners and pipelining make up implementation of the underlying algorithms. Moreover, we tried to design the FACET API to align with the scikit-learn API. - The
SHAP <https://shap.readthedocs.io/en/latest/>__ implementation is used to estimate the shapley vectors which FACET then decomposes into synergy, redundancy, and independence vectors.
If you would like to know more about the team behind FACET please see the
about us <https://bcg-x-official.github.io/facet/about_us.html>__ page.
We are always on the lookout for passionate and talented data scientists to join the
BCG GAMMA team. If you would like to know more you can find out about
BCG GAMMA <https://www.bcg.com/en-gb/beyond-consulting/bcg-gamma/default>,
or have a look at
career opportunities <https://www.bcg.com/en-gb/beyond-consulting/bcg-gamma/careers>.
.. |pipe| image:: sphinx/source/_images/icons/pipe_icon.png :width: 100px :class: facet_icon
.. |inspect| image:: sphinx/source/_images/icons/inspect_icon.png :width: 100px :class: facet_icon
.. |sim| image:: sphinx/source/_images/icons/sim_icon.png :width: 100px :class: facet_icon
.. |spacer| unicode:: 0x2003 0x2003 0x2003 0x2003 0x2003 0x2003
.. Begin-Badges
.. |conda| image:: https://anaconda.org/bcg_gamma/gamma-facet/badges/version.svg :target: https://anaconda.org/BCG_Gamma/gamma-facet
.. |pypi| image:: https://badge.fury.io/py/gamma-facet.svg :target: https://pypi.org/project/gamma-facet/
.. |azure_build| image:: https://dev.azure.com/gamma-facet/facet/_apis/build/status/BCG-X-Official.facet?repoName=BCG-X-Official%2Ffacet&branchName=develop :target: https://dev.azure.com/gamma-facet/facet/_build?definitionId=7&_a=summary
.. |azure_code_cov| image:: https://img.shields.io/azure-devops/coverage/gamma-facet/facet/7/2.0.x :target: https://dev.azure.com/gamma-facet/facet/_build?definitionId=7&_a=summary
.. |python_versions| image:: https://img.shields.io/badge/python-3.7|3.8|3.9-blue.svg :target: https://www.python.org/downloads/release/python-380/
.. |code_style| image:: https://img.shields.io/badge/code%20style-black-000000.svg :target: https://github.com/psf/black
.. |made_with_sphinx_doc| image:: https://img.shields.io/badge/Made%20with-Sphinx-1f425f.svg :target: https://bcg-x-official.github.io/facet/index.html
.. |license_badge| image:: https://img.shields.io/badge/License-Apache%202.0-olivegreen.svg :target: https://opensource.org/licenses/Apache-2.0
.. End-Badges
For Tasks:
Click tags to check more tools for each tasksFor Jobs:
Alternative AI tools for facet
Similar Open Source Tools

facet
FACET is an open source library for human-explainable AI that combines model inspection and model-based simulation to provide better explanations for supervised machine learning models. It offers an efficient and transparent machine learning workflow, enhancing scikit-learn's pipelining paradigm with new capabilities for model selection, inspection, and simulation. FACET introduces new algorithms for quantifying dependencies and interactions between features in ML models, as well as for conducting virtual experiments to optimize predicted outcomes. The tool ensures end-to-end traceability of features using an augmented version of scikit-learn with enhanced support for pandas data frames. FACET also provides model inspection methods for scikit-learn estimators, enhancing global metrics like synergy and redundancy to complement the local perspective of SHAP. Additionally, FACET offers model simulation capabilities for conducting univariate uplift simulations based on important features like BMI.
llms
The 'llms' repository is a comprehensive guide on Large Language Models (LLMs), covering topics such as language modeling, applications of LLMs, statistical language modeling, neural language models, conditional language models, evaluation methods, transformer-based language models, practical LLMs like GPT and BERT, prompt engineering, fine-tuning LLMs, retrieval augmented generation, AI agents, and LLMs for computer vision. The repository provides detailed explanations, examples, and tools for working with LLMs.
gepa
GEPA (Genetic-Pareto) is a framework for optimizing arbitrary systems composed of text components like AI prompts, code snippets, or textual specs against any evaluation metric. It employs LLMs to reflect on system behavior, using feedback from execution and evaluation traces to drive targeted improvements. Through iterative mutation, reflection, and Pareto-aware candidate selection, GEPA evolves robust, high-performing variants with minimal evaluations, co-evolving multiple components in modular systems for domain-specific gains. The repository provides the official implementation of the GEPA algorithm as proposed in the paper titled 'GEPA: Reflective Prompt Evolution Can Outperform Reinforcement Learning'.

zshot
Zshot is a highly customizable framework for performing Zero and Few shot named entity and relationships recognition. It can be used for mentions extraction, wikification, zero and few shot named entity recognition, zero and few shot named relationship recognition, and visualization of zero-shot NER and RE extraction. The framework consists of two main components: the mentions extractor and the linker. There are multiple mentions extractors and linkers available, each serving a specific purpose. Zshot also includes a relations extractor and a knowledge extractor for extracting relations among entities and performing entity classification. The tool requires Python 3.6+ and dependencies like spacy, torch, transformers, evaluate, and datasets for evaluation over datasets like OntoNotes. Optional dependencies include flair and blink for additional functionalities. Zshot provides examples, tutorials, and evaluation methods to assess the performance of the components.
kafka-ml
Kafka-ML is a framework designed to manage the pipeline of Tensorflow/Keras and PyTorch machine learning models on Kubernetes. It enables the design, training, and inference of ML models with datasets fed through Apache Kafka, connecting them directly to data streams like those from IoT devices. The Web UI allows easy definition of ML models without external libraries, catering to both experts and non-experts in ML/AI.
qlib
Qlib is an open-source, AI-oriented quantitative investment platform that supports diverse machine learning modeling paradigms, including supervised learning, market dynamics modeling, and reinforcement learning. It covers the entire chain of quantitative investment, from alpha seeking to order execution. The platform empowers researchers to explore ideas and implement productions using AI technologies in quantitative investment. Qlib collaboratively solves key challenges in quantitative investment by releasing state-of-the-art research works in various paradigms. It provides a full ML pipeline for data processing, model training, and back-testing, enabling users to perform tasks such as forecasting market patterns, adapting to market dynamics, and modeling continuous investment decisions.
RLHF-Reward-Modeling
This repository contains code for training reward models for Deep Reinforcement Learning-based Reward-modulated Hierarchical Fine-tuning (DRL-based RLHF), Iterative Selection Fine-tuning (Rejection sampling fine-tuning), and iterative Decision Policy Optimization (DPO). The reward models are trained using a Bradley-Terry model based on the Gemma and Mistral language models. The resulting reward models achieve state-of-the-art performance on the RewardBench leaderboard for reward models with base models of up to 13B parameters.
fortuna
Fortuna is a library for uncertainty quantification that enables users to estimate predictive uncertainty, assess model reliability, trigger human intervention, and deploy models safely. It provides calibration and conformal methods for pre-trained models in any framework, supports Bayesian inference methods for deep learning models written in Flax, and is designed to be intuitive and highly configurable. Users can run benchmarks and bring uncertainty to production systems with ease.
uncheatable_eval
Uncheatable Eval is a tool designed to assess the language modeling capabilities of LLMs on real-time, newly generated data from the internet. It aims to provide a reliable evaluation method that is immune to data leaks and cannot be gamed. The tool supports the evaluation of Hugging Face AutoModelForCausalLM models and RWKV models by calculating the sum of negative log probabilities on new texts from various sources such as recent papers on arXiv, new projects on GitHub, news articles, and more. Uncheatable Eval ensures that the evaluation data is not included in the training sets of publicly released models, thus offering a fair assessment of the models' performance.

llm-reasoners
LLM Reasoners is a library that enables LLMs to conduct complex reasoning, with advanced reasoning algorithms. It approaches multi-step reasoning as planning and searches for the optimal reasoning chain, which achieves the best balance of exploration vs exploitation with the idea of "World Model" and "Reward". Given any reasoning problem, simply define the reward function and an optional world model (explained below), and let LLM reasoners take care of the rest, including Reasoning Algorithms, Visualization, LLM calling, and more!

OneKE
OneKE is a flexible dockerized system for schema-guided knowledge extraction, capable of extracting information from the web and raw PDF books across multiple domains like science and news. It employs a collaborative multi-agent approach and includes a user-customizable knowledge base to enable tailored extraction. OneKE offers various IE tasks support, data sources support, LLMs support, extraction method support, and knowledge base configuration. Users can start with examples using YAML, Python, or Web UI, and perform tasks like Named Entity Recognition, Relation Extraction, Event Extraction, Triple Extraction, and Open Domain IE. The tool supports different source formats like Plain Text, HTML, PDF, Word, TXT, and JSON files. Users can choose from various extraction models like OpenAI, DeepSeek, LLaMA, Qwen, ChatGLM, MiniCPM, and OneKE for information extraction tasks. Extraction methods include Schema Agent, Extraction Agent, and Reflection Agent. The tool also provides support for schema repository and case repository management, along with solutions for network issues. Contributors to the project include Ningyu Zhang, Haofen Wang, Yujie Luo, Xiangyuan Ru, Kangwei Liu, Lin Yuan, Mengshu Sun, Lei Liang, Zhiqiang Zhang, Jun Zhou, Lanning Wei, Da Zheng, and Huajun Chen.
judges
The 'judges' repository is a small library designed for using and creating LLM-as-a-Judge evaluators. It offers a curated set of LLM evaluators in a low-friction format for various use cases, backed by research. Users can use these evaluators off-the-shelf or as inspiration for building custom LLM evaluators. The library provides two types of judges: Classifiers that return boolean values and Graders that return scores on a numerical or Likert scale. Users can combine multiple judges using the 'Jury' object and evaluate input-output pairs with the '.judge()' method. Additionally, the repository includes detailed instructions on picking a model, sending data to an LLM, using classifiers, combining judges, and creating custom LLM judges with 'AutoJudge'.
storm
STORM is a LLM system that writes Wikipedia-like articles from scratch based on Internet search. While the system cannot produce publication-ready articles that often require a significant number of edits, experienced Wikipedia editors have found it helpful in their pre-writing stage. **Try out our [live research preview](https://storm.genie.stanford.edu/) to see how STORM can help your knowledge exploration journey and please provide feedback to help us improve the system 🙏!**
Quantus
Quantus is a toolkit designed for the evaluation of neural network explanations. It offers more than 30 metrics in 6 categories for eXplainable Artificial Intelligence (XAI) evaluation. The toolkit supports different data types (image, time-series, tabular, NLP) and models (PyTorch, TensorFlow). It provides built-in support for explanation methods like captum, tf-explain, and zennit. Quantus is under active development and aims to provide a comprehensive set of quantitative evaluation metrics for XAI methods.
OREAL
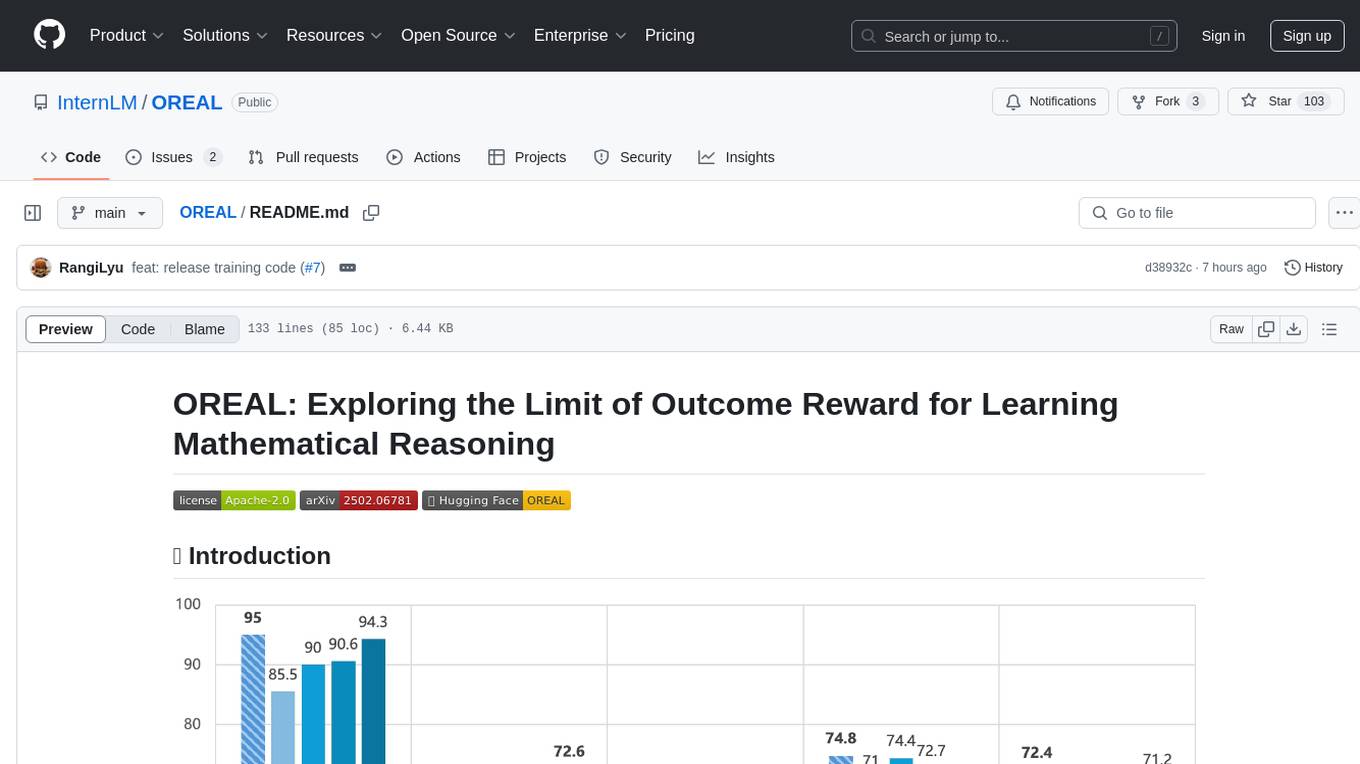
OREAL is a reinforcement learning framework designed for mathematical reasoning tasks, aiming to achieve optimal performance through outcome reward-based learning. The framework utilizes behavior cloning, reshaping rewards, and token-level reward models to address challenges in sparse rewards and partial correctness. OREAL has achieved significant results, with a 7B model reaching 94.0 pass@1 accuracy on MATH-500 and surpassing previous 32B models. The tool provides training tutorials and Hugging Face model repositories for easy access and implementation.
pint-benchmark
The Lakera PINT Benchmark provides a neutral evaluation method for prompt injection detection systems, offering a dataset of English inputs with prompt injections, jailbreaks, benign inputs, user-agent chats, and public document excerpts. The dataset is designed to be challenging and representative, with plans for future enhancements. The benchmark aims to be unbiased and accurate, welcoming contributions to improve prompt injection detection. Users can evaluate prompt injection detection systems using the provided Jupyter Notebook. The dataset structure is specified in YAML format, allowing users to prepare their datasets for benchmarking. Evaluation examples and resources are provided to assist users in evaluating prompt injection detection models and tools.
For similar tasks

facet
FACET is an open source library for human-explainable AI that combines model inspection and model-based simulation to provide better explanations for supervised machine learning models. It offers an efficient and transparent machine learning workflow, enhancing scikit-learn's pipelining paradigm with new capabilities for model selection, inspection, and simulation. FACET introduces new algorithms for quantifying dependencies and interactions between features in ML models, as well as for conducting virtual experiments to optimize predicted outcomes. The tool ensures end-to-end traceability of features using an augmented version of scikit-learn with enhanced support for pandas data frames. FACET also provides model inspection methods for scikit-learn estimators, enhancing global metrics like synergy and redundancy to complement the local perspective of SHAP. Additionally, FACET offers model simulation capabilities for conducting univariate uplift simulations based on important features like BMI.
Detection-and-Classification-of-Alzheimers-Disease
This tool is designed to detect and classify Alzheimer's Disease using Deep Learning and Machine Learning algorithms on an early basis, which is further optimized using the Crow Search Algorithm (CSA). Alzheimer's is a fatal disease, and early detection is crucial for patients to predetermine their condition and prevent its progression. By analyzing MRI scanned images using Artificial Intelligence technology, this tool can classify patients who may or may not develop AD in the future. The CSA algorithm, combined with ML algorithms, has proven to be the most effective approach for this purpose.
For similar jobs
weave
Weave is a toolkit for developing Generative AI applications, built by Weights & Biases. With Weave, you can log and debug language model inputs, outputs, and traces; build rigorous, apples-to-apples evaluations for language model use cases; and organize all the information generated across the LLM workflow, from experimentation to evaluations to production. Weave aims to bring rigor, best-practices, and composability to the inherently experimental process of developing Generative AI software, without introducing cognitive overhead.

LLMStack
LLMStack is a no-code platform for building generative AI agents, workflows, and chatbots. It allows users to connect their own data, internal tools, and GPT-powered models without any coding experience. LLMStack can be deployed to the cloud or on-premise and can be accessed via HTTP API or triggered from Slack or Discord.

VisionCraft
The VisionCraft API is a free API for using over 100 different AI models. From images to sound.
kaito
Kaito is an operator that automates the AI/ML inference model deployment in a Kubernetes cluster. It manages large model files using container images, avoids tuning deployment parameters to fit GPU hardware by providing preset configurations, auto-provisions GPU nodes based on model requirements, and hosts large model images in the public Microsoft Container Registry (MCR) if the license allows. Using Kaito, the workflow of onboarding large AI inference models in Kubernetes is largely simplified.

PyRIT
PyRIT is an open access automation framework designed to empower security professionals and ML engineers to red team foundation models and their applications. It automates AI Red Teaming tasks to allow operators to focus on more complicated and time-consuming tasks and can also identify security harms such as misuse (e.g., malware generation, jailbreaking), and privacy harms (e.g., identity theft). The goal is to allow researchers to have a baseline of how well their model and entire inference pipeline is doing against different harm categories and to be able to compare that baseline to future iterations of their model. This allows them to have empirical data on how well their model is doing today, and detect any degradation of performance based on future improvements.
tabby
Tabby is a self-hosted AI coding assistant, offering an open-source and on-premises alternative to GitHub Copilot. It boasts several key features: * Self-contained, with no need for a DBMS or cloud service. * OpenAPI interface, easy to integrate with existing infrastructure (e.g Cloud IDE). * Supports consumer-grade GPUs.
spear
SPEAR (Simulator for Photorealistic Embodied AI Research) is a powerful tool for training embodied agents. It features 300 unique virtual indoor environments with 2,566 unique rooms and 17,234 unique objects that can be manipulated individually. Each environment is designed by a professional artist and features detailed geometry, photorealistic materials, and a unique floor plan and object layout. SPEAR is implemented as Unreal Engine assets and provides an OpenAI Gym interface for interacting with the environments via Python.
Magick
Magick is a groundbreaking visual AIDE (Artificial Intelligence Development Environment) for no-code data pipelines and multimodal agents. Magick can connect to other services and comes with nodes and templates well-suited for intelligent agents, chatbots, complex reasoning systems and realistic characters.