TxAgent
TxAgent: An AI Agent for Therapeutic Reasoning Across a Universe of Tools
Stars: 382
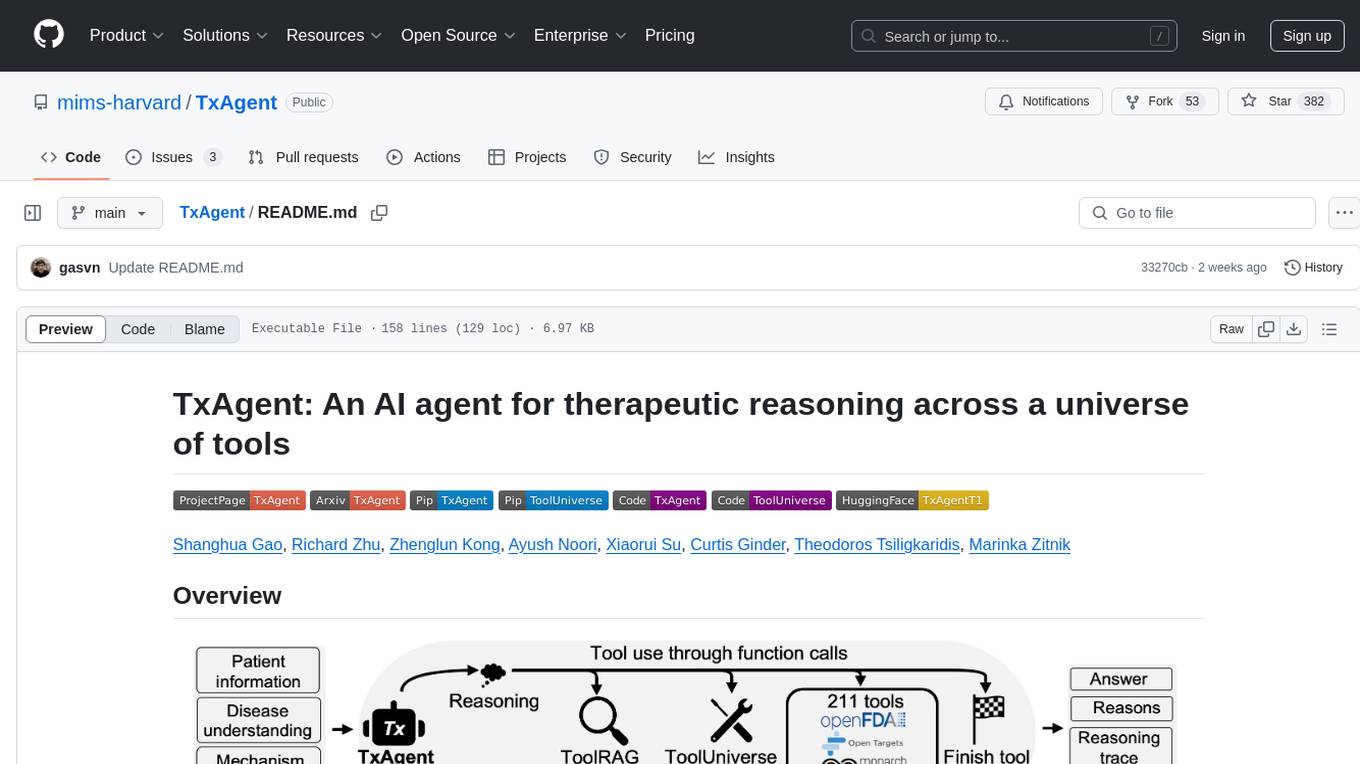
TxAgent is an AI agent designed for precision therapeutics, leveraging multi-step reasoning and real-time biomedical knowledge retrieval across a toolbox of 211 tools. It evaluates drug interactions, contraindications, and tailors treatment strategies to individual patient characteristics. TxAgent outperforms leading models across various drug reasoning tasks and personalized treatment scenarios, ensuring treatment recommendations align with clinical guidelines and real-world evidence.
README:
Precision therapeutics require multimodal adaptive models that generate personalized treatment recommendations. We introduce TxAgent, an AI agent that leverages multi-step reasoning and real-time biomedical knowledge retrieval across a toolbox of 211 tools to analyze drug interactions, contraindications, and patient-specific treatment strategies.
- TxAgent evaluates how drugs interact at molecular, pharmacokinetic, and clinical levels, identifies contraindications based on patient comorbidities and concurrent medications, and tailors treatment strategies to individual patient characteristics, including age, genetic factors, and disease progression.
- TxAgent retrieves and synthesizes evidence from multiple biomedical sources, assesses interactions between drugs and patient conditions, and refines treatment recommendations through iterative reasoning. It selects tools based on task objectives and executes structured function calls to solve therapeutic tasks that require clinical reasoning and cross-source validation.
- The ToolUniverse consolidates 211 tools from trusted sources, including all US FDA-approved drugs since 1939 and validated clinical insights from Open Targets.
TxAgent outperforms leading LLMs, tool-use models, and reasoning agents across five new benchmarks: DrugPC, BrandPC, GenericPC, TreatmentPC, and DescriptionPC, covering 3,168 drug reasoning tasks and 456 personalized treatment scenarios.
- It achieves 92.1% accuracy in open-ended drug reasoning tasks, surpassing GPT-4o by up to 25.8% and outperforming DeepSeek-R1 (671B) in structured multi-step reasoning.
- TxAgent generalizes across drug name variants and descriptions, maintaining a variance of < 0.01 between brand, generic, and description-based drug references, exceeding existing tool-use LLMs by over 55%.
By integrating multi-step inference, real-time knowledge grounding, and tool- assisted decision-making, TxAgent ensures that treatment recommendations align with established clinical guidelines and real-world evidence, reducing the risk of adverse events and improving therapeutic decision-making.
Dependency:
- An H100 GPU with more than 80GB of memory is recommended when running TxAgent.
- ToolUniverse requires a device with an internet connection.
Install ToolUniverse:
# Install from source code:
git clone https://github.com/mims-harvard/ToolUniverse.git
cd ToolUniverse
python -m pip install . --no-cache-dir
OR
# Install from pip:
pip install tooluniverse
Install TxAgent:
# Install from source code:
git clone https://github.com/mims-harvard/TxAgent.git
python -m pip install . --no-cache-dir
OR
# Install from pip:
pip install txagent
Run the example:
python run_example.py
Run the gradio demo:
python run_txagent_app.py
Pretrained model weights are available in HuggingFace.
| Model | Description |
|---|---|
| TxAgent-T1-Llama-3.1-8B | TxAgent LLM |
| ToolRAG-T1-GTE-Qwen2-1.5B | Tool RAG embedding model |
Please visit project page for more details.



@misc{gao2025txagent,
title={TxAgent: An AI Agent for Therapeutic Reasoning Across a Universe of Tools},
author={Shanghua Gao and Richard Zhu and Zhenglun Kong and Ayush Noori and Xiaorui Su and Curtis Ginder and Theodoros Tsiligkaridis and Marinka Zitnik},
year={2025},
eprint={2503.10970},
archivePrefix={arXiv},
primaryClass={cs.AI},
url={https://arxiv.org/abs/2503.10970},
}
If you have any questions or suggestions, please email Shanghua Gao and Marinka Zitnik.
For Tasks:
Click tags to check more tools for each tasksFor Jobs:
Alternative AI tools for TxAgent
Similar Open Source Tools

TxAgent
TxAgent is an AI agent designed for precision therapeutics, leveraging multi-step reasoning and real-time biomedical knowledge retrieval across a toolbox of 211 tools. It evaluates drug interactions, contraindications, and tailors treatment strategies to individual patient characteristics. TxAgent outperforms leading models across various drug reasoning tasks and personalized treatment scenarios, ensuring treatment recommendations align with clinical guidelines and real-world evidence.
RD-Agent
RD-Agent is a tool designed to automate critical aspects of industrial R&D processes, focusing on data-driven scenarios to streamline model and data development. It aims to propose new ideas ('R') and implement them ('D') automatically, leading to solutions of significant industrial value. The tool supports scenarios like Automated Quantitative Trading, Data Mining Agent, Research Copilot, and more, with a framework to push the boundaries of research in data science. Users can create a Conda environment, install the RDAgent package from PyPI, configure GPT model, and run various applications for tasks like quantitative trading, model evolution, medical prediction, and more. The tool is intended to enhance R&D processes and boost productivity in industrial settings.
FuseAI
FuseAI is a repository that focuses on knowledge fusion of large language models. It includes FuseChat, a state-of-the-art 7B LLM on MT-Bench, and FuseLLM, which surpasses Llama-2-7B by fusing three open-source foundation LLMs. The repository provides tech reports, releases, and datasets for FuseChat and FuseLLM, showcasing their performance and advancements in the field of chat models and large language models.

LightLLM
LightLLM is a lightweight library for linear and logistic regression models. It provides a simple and efficient way to train and deploy machine learning models for regression tasks. The library is designed to be easy to use and integrate into existing projects, making it suitable for both beginners and experienced data scientists. With LightLLM, users can quickly build and evaluate regression models using a variety of algorithms and hyperparameters. The library also supports feature engineering and model interpretation, allowing users to gain insights from their data and make informed decisions based on the model predictions.
CuMo
CuMo is a project focused on scaling multimodal Large Language Models (LLMs) with Co-Upcycled Mixture-of-Experts. It introduces CuMo, which incorporates Co-upcycled Top-K sparsely-gated Mixture-of-experts blocks into the vision encoder and the MLP connector, enhancing the capabilities of multimodal LLMs. The project adopts a three-stage training approach with auxiliary losses to stabilize the training process and maintain a balanced loading of experts. CuMo achieves comparable performance to other state-of-the-art multimodal LLMs on various Visual Question Answering (VQA) and visual-instruction-following benchmarks.

lerobot
LeRobot is a state-of-the-art AI library for real-world robotics in PyTorch. It aims to provide models, datasets, and tools to lower the barrier to entry to robotics, focusing on imitation learning and reinforcement learning. LeRobot offers pretrained models, datasets with human-collected demonstrations, and simulation environments. It plans to support real-world robotics on affordable and capable robots. The library hosts pretrained models and datasets on the Hugging Face community page.

AdalFlow
AdalFlow is a library designed to help developers build and optimize Large Language Model (LLM) task pipelines. It follows a design pattern similar to PyTorch, offering a light, modular, and robust codebase. Named in honor of Ada Lovelace, AdalFlow aims to inspire more women to enter the AI field. The library is tailored for various GenAI applications like chatbots, translation, summarization, code generation, and autonomous agents, as well as classical NLP tasks such as text classification and named entity recognition. AdalFlow emphasizes modularity, robustness, and readability to support users in customizing and iterating code for their specific use cases.
pytorch-forecasting
PyTorch Forecasting is a PyTorch-based package designed for state-of-the-art timeseries forecasting using deep learning architectures. It offers a high-level API and leverages PyTorch Lightning for efficient training on GPU or CPU with automatic logging. The package aims to simplify timeseries forecasting tasks by providing a flexible API for professionals and user-friendly defaults for beginners. It includes features such as a timeseries dataset class for handling data transformations, missing values, and subsampling, various neural network architectures optimized for real-world deployment, multi-horizon timeseries metrics, and hyperparameter tuning with optuna. Built on pytorch-lightning, it supports training on CPUs, single GPUs, and multiple GPUs out-of-the-box.

openrl
OpenRL is an open-source general reinforcement learning research framework that supports training for various tasks such as single-agent, multi-agent, offline RL, self-play, and natural language. Developed based on PyTorch, the goal of OpenRL is to provide a simple-to-use, flexible, efficient and sustainable platform for the reinforcement learning research community. It supports a universal interface for all tasks/environments, single-agent and multi-agent tasks, offline RL training with expert dataset, self-play training, reinforcement learning training for natural language tasks, DeepSpeed, Arena for evaluation, importing models and datasets from Hugging Face, user-defined environments, models, and datasets, gymnasium environments, callbacks, visualization tools, unit testing, and code coverage testing. It also supports various algorithms like PPO, DQN, SAC, and environments like Gymnasium, MuJoCo, Atari, and more.
lobe-chat
Lobe Chat is an open-source, modern-design ChatGPT/LLMs UI/Framework. Supports speech-synthesis, multi-modal, and extensible ([function call][docs-functionc-call]) plugin system. One-click **FREE** deployment of your private OpenAI ChatGPT/Claude/Gemini/Groq/Ollama chat application.
qgate-model
QGate-Model is a machine learning meta-model with synthetic data, designed for MLOps and feature store. It is independent of machine learning solutions, with definitions in JSON and data in CSV/parquet formats. This meta-model is useful for comparing capabilities and functions of machine learning solutions, independently testing new versions of machine learning solutions, and conducting various types of tests (unit, sanity, smoke, system, regression, function, acceptance, performance, shadow, etc.). It can also be used for external test coverage when internal test coverage is not available or weak.
OmAgent
OmAgent is an open-source agent framework designed to streamline the development of on-device multimodal agents. It enables agents to empower various hardware devices, integrates speed-optimized SOTA multimodal models, provides SOTA multimodal agent algorithms, and focuses on optimizing the end-to-end computing pipeline for real-time user interaction experience. Key features include easy connection to diverse devices, scalability, flexibility, and workflow orchestration. The architecture emphasizes graph-based workflow orchestration, native multimodality, and device-centricity, allowing developers to create bespoke intelligent agent programs.
agents-towards-production
Agents Towards Production is an open-source playbook for building production-ready GenAI agents that scale from prototype to enterprise. Tutorials cover stateful workflows, vector memory, real-time web search APIs, Docker deployment, FastAPI endpoints, security guardrails, GPU scaling, browser automation, fine-tuning, multi-agent coordination, observability, evaluation, and UI development.
ludwig
Ludwig is a declarative deep learning framework designed for scale and efficiency. It is a low-code framework that allows users to build custom AI models like LLMs and other deep neural networks with ease. Ludwig offers features such as optimized scale and efficiency, expert level control, modularity, and extensibility. It is engineered for production with prebuilt Docker containers, support for running with Ray on Kubernetes, and the ability to export models to Torchscript and Triton. Ludwig is hosted by the Linux Foundation AI & Data.
AceCoder
AceCoder is a tool that introduces a fully automated pipeline for synthesizing large-scale reliable tests used for reward model training and reinforcement learning in the coding scenario. It curates datasets, trains reward models, and performs RL training to improve coding abilities of language models. The tool aims to unlock the potential of RL training for code generation models and push the boundaries of LLM's coding abilities.
For similar tasks

TxAgent
TxAgent is an AI agent designed for precision therapeutics, leveraging multi-step reasoning and real-time biomedical knowledge retrieval across a toolbox of 211 tools. It evaluates drug interactions, contraindications, and tailors treatment strategies to individual patient characteristics. TxAgent outperforms leading models across various drug reasoning tasks and personalized treatment scenarios, ensuring treatment recommendations align with clinical guidelines and real-world evidence.
For similar jobs

KG_RAG
KG-RAG (Knowledge Graph-based Retrieval Augmented Generation) is a task agnostic framework that combines the explicit knowledge of a Knowledge Graph (KG) with the implicit knowledge of a Large Language Model (LLM). KG-RAG extracts "prompt-aware context" from a KG, which is defined as the minimal context sufficient enough to respond to the user prompt. This framework empowers a general-purpose LLM by incorporating an optimized domain-specific 'prompt-aware context' from a biomedical KG. KG-RAG is specifically designed for running prompts related to Diseases.

Scientific-LLM-Survey
Scientific Large Language Models (Sci-LLMs) is a repository that collects papers on scientific large language models, focusing on biology and chemistry domains. It includes textual, molecular, protein, and genomic languages, as well as multimodal language. The repository covers various large language models for tasks such as molecule property prediction, interaction prediction, protein sequence representation, protein sequence generation/design, DNA-protein interaction prediction, and RNA prediction. It also provides datasets and benchmarks for evaluating these models. The repository aims to facilitate research and development in the field of scientific language modeling.
biochatter
Generative AI models have shown tremendous usefulness in increasing accessibility and automation of a wide range of tasks. This repository contains the `biochatter` Python package, a generic backend library for the connection of biomedical applications to conversational AI. It aims to provide a common framework for deploying, testing, and evaluating diverse models and auxiliary technologies in the biomedical domain. BioChatter is part of the BioCypher ecosystem, connecting natively to BioCypher knowledge graphs.
cellseg_models.pytorch
cellseg-models.pytorch is a Python library built upon PyTorch for 2D cell/nuclei instance segmentation models. It provides multi-task encoder-decoder architectures and post-processing methods for segmenting cell/nuclei instances. The library offers high-level API to define segmentation models, open-source datasets for training, flexibility to modify model components, sliding window inference, multi-GPU inference, benchmarking utilities, regularization techniques, and example notebooks for training and finetuning models with different backbones.
aicsimageio
AICSImageIO is a Python tool for Image Reading, Metadata Conversion, and Image Writing for Microscopy Images. It supports various file formats like OME-TIFF, TIFF, ND2, DV, CZI, LIF, PNG, GIF, and Bio-Formats. Users can read and write metadata and imaging data, work with different file systems like local paths, HTTP URLs, s3fs, and gcsfs. The tool provides functionalities for full image reading, delayed image reading, mosaic image reading, metadata reading, xarray coordinate plane attachment, cloud IO support, and saving to OME-TIFF. It also offers benchmarking and developer resources.
ceLLama
ceLLama is a streamlined automation pipeline for cell type annotations using large-language models (LLMs). It operates locally to ensure privacy, provides comprehensive analysis by considering negative genes, offers efficient processing speed, and generates customized reports. Ideal for quick and preliminary cell type checks.
PINNACLE
PINNACLE is a flexible geometric deep learning approach that trains on contextualized protein interaction networks to generate context-aware protein representations. It provides protein representations split across various cell-type contexts from different tissues and organs. The tool can be fine-tuned to study the genomic effects of drugs and nominate promising protein targets and cell-type contexts for further investigation. PINNACLE exemplifies the paradigm of incorporating context-specific effects for studying biological systems, especially the impact of disease and therapeutics.
Taiyi-LLM
Taiyi (太一) is a bilingual large language model fine-tuned for diverse biomedical tasks. It aims to facilitate communication between healthcare professionals and patients, provide medical information, and assist in diagnosis, biomedical knowledge discovery, drug development, and personalized healthcare solutions. The model is based on the Qwen-7B-base model and has been fine-tuned using rich bilingual instruction data. It covers tasks such as question answering, biomedical dialogue, medical report generation, biomedical information extraction, machine translation, title generation, text classification, and text semantic similarity. The project also provides standardized data formats, model training details, model inference guidelines, and overall performance metrics across various BioNLP tasks.