Best AI tools for< Computational Chemist >
Infographic
20 - AI tool Sites

XtalPi
XtalPi is a world-leading technology company driven by artificial intelligence (AI) and robotics to innovate in the fields of life sciences and new materials. Founded in 2015 at the Massachusetts Institute of Technology (MIT), the company is committed to realizing digital and intelligent innovation in the fields of life sciences and new materials. Based on cutting-edge technologies and capabilities such as quantum physics, artificial intelligence, cloud computing, and large-scale experimental robot clusters, the company provides innovative technologies, services, and products for global industries such as biomedicine, chemicals, new energy, and new materials.
Iambic Therapeutics
Iambic Therapeutics is a cutting-edge AI-driven drug discovery platform that tackles the most challenging design problems in drug discovery, addressing unmet patient need. Its physics-based AI algorithms drive a high-throughput experimental platform, converting new molecular designs to new biological insights each week. Iambic's platform optimizes target product profiles, exploring multiple profiles in parallel to ensure that molecules are designed to solve the right problems in disease biology. It also optimizes drug candidates, deeply exploring chemical space to reveal novel mechanisms of action and deliver diverse high-quality leads.
Variational AI
Variational AI is a company that uses generative AI to discover novel drug-like small molecules with optimized properties for defined targets. Their platform, Enki™, is the first commercially accessible foundation model for small molecules. It is designed to make generating novel molecule structures easy, with no data required. Users simply define their target product profile (TPP) and Enki does the rest. Enki is an ensemble of generative algorithms trained on decades worth of experimental data with proven results. The company was founded in September 2019 and is based in Vancouver, BC, Canada.

Allchemy
Allchemy is a resource-aware AI platform for drug discovery. It combines state-of-the-art computational synthesis with AI algorithms to predict molecular properties. Within minutes, Allchemy creates thousands of synthesizable lead candidates meeting user-defined profiles of drug-likeness, affinity towards specific proteins, toxicity, and a range of other physical-chemical measures. Allchemy encompasses the entire resource-to-drug design process and has been used in academic, corporate and classified environments worldwide to: Design synthesizable leads targeting specific proteins Evolve scaffolds similar to desired drugs Design “circular” drug syntheses from renewable materials Interface with and instruct automated synthesis platforms and optimize pilot-scale processes Operate “iterative synthesis” schemes Predict side reactions and create forensic “synthetic signatures” of hazardous/toxic molecules Design synthetic degradation and recovery cycles for various types of feedstocks and functional target molecules
Lavo Life Sciences
Lavo Life Sciences is an AI-accelerated crystal structure prediction application that helps in drug development by providing accurate predictions for small molecule drugs. The application utilizes AI technology to optimize solid-state formulations, reduce turnaround time, mitigate risks, and discover novel polymorphs, ultimately streamlining the pharmaceutical research and development process.
Cradle
Cradle is a protein engineering platform that uses machine learning to design improved protein sequences. It allows users to import assay data, generate new sequences, test them in the lab, and import the results to improve the model. Cradle can be used to optimize multiple properties of a protein simultaneously, and it has been used by leading biotech teams to accelerate new and ongoing projects.
Genesis Therapeutics
Genesis Therapeutics is an AI platform that leverages cutting-edge technology to revolutionize drug discovery and development processes. The platform integrates advanced algorithms and machine learning models to accelerate the identification of novel drug candidates and optimize their properties. By combining computational simulations with experimental data, Genesis Therapeutics offers a comprehensive solution to streamline the drug development pipeline and bring innovative therapies to market faster. The platform is designed to empower researchers and pharmaceutical companies with powerful tools for predicting drug-target interactions, optimizing molecular structures, and prioritizing lead compounds for further investigation.
Institute for Protein Design
The Institute for Protein Design is a research institute at the University of Washington that uses computational design to create new proteins that solve modern challenges in medicine, technology, and sustainability. The institute's research focuses on developing new protein therapeutics, vaccines, drug delivery systems, biological devices, self-assembling nanomaterials, and bioactive peptides. The institute also has a strong commitment to responsible AI development and has developed a set of principles to guide its use of AI in research.

Julius AI
Julius AI is an advanced AI data analyst tool that allows users to analyze data with computational AI, chat with files to get expert-level insights, create sleek data visualizations, perform modeling and predictive forecasting, solve math, physics, and chemistry problems, generate polished analyses and summaries, save time by automating data work, and unlock statistical modeling without complexity. It offers features like generating visualizations, asking data questions, effortless cleaning, instant data export, creating animations, and supercharging data analysis. Julius AI is loved by over 1,200,000 users worldwide and is designed to help knowledge workers make the most out of their data.

Artificial Intelligence: Foundations of Computational Agents
Artificial Intelligence: Foundations of Computational Agents, 3rd edition by David L. Poole and Alan K. Mackworth, Cambridge University Press 2023, is a book about the science of artificial intelligence (AI). It presents artificial intelligence as the study of the design of intelligent computational agents. The book is structured as a textbook, but it is accessible to a wide audience of professionals and researchers. In the last decades we have witnessed the emergence of artificial intelligence as a serious science and engineering discipline. This book provides an accessible synthesis of the field aimed at undergraduate and graduate students. It provides a coherent vision of the foundations of the field as it is today. It aims to provide that synthesis as an integrated science, in terms of a multi-dimensional design space that has been partially explored. As with any science worth its salt, artificial intelligence has a coherent, formal theory and a rambunctious experimental wing. The book balances theory and experiment, showing how to link them intimately together. It develops the science of AI together with its engineering applications.
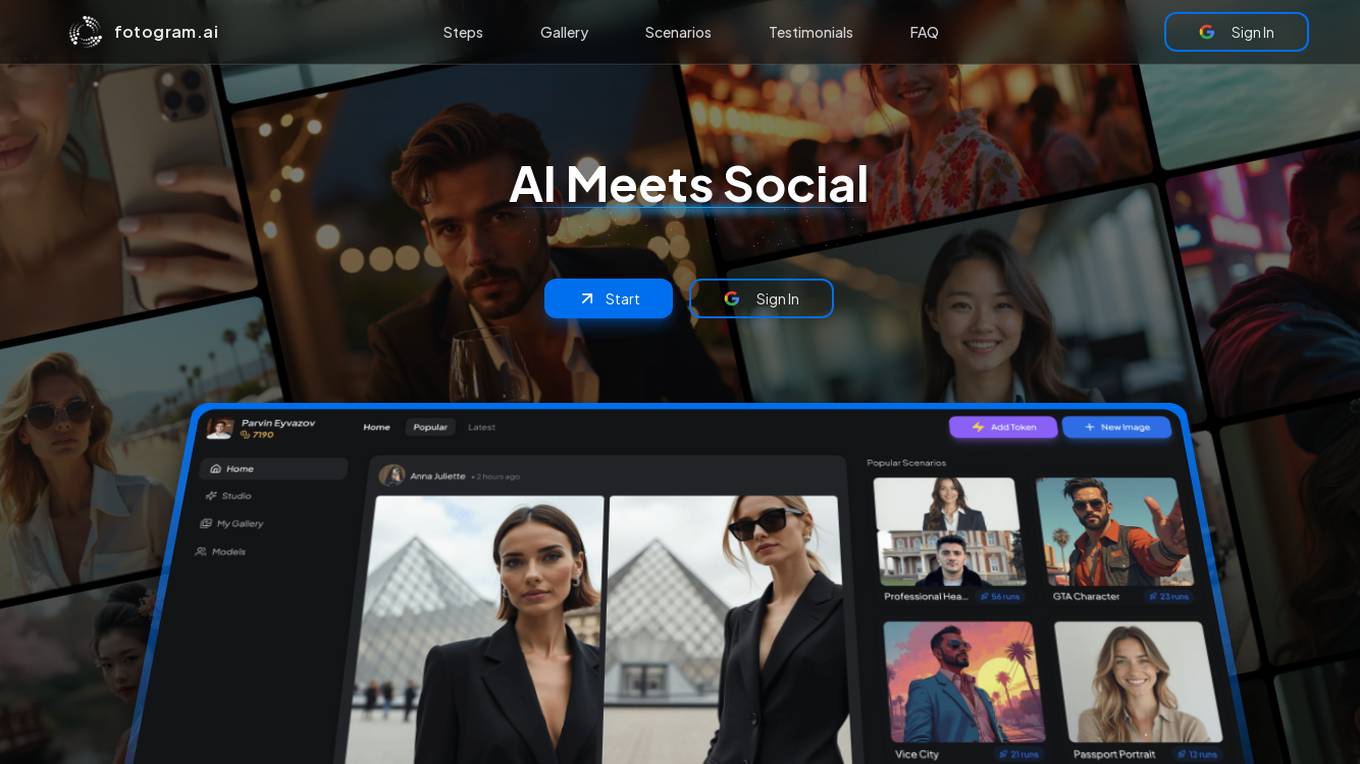
Fotogram.ai
Fotogram.ai is an AI-powered image editing tool that offers a wide range of features to enhance and transform your photos. With Fotogram.ai, users can easily apply filters, adjust colors, remove backgrounds, add effects, and retouch images with just a few clicks. The tool uses advanced AI algorithms to provide professional-level editing capabilities to users of all skill levels. Whether you are a photographer looking to streamline your workflow or a social media enthusiast wanting to create stunning visuals, Fotogram.ai has you covered.
PAACADEMY
PAACADEMY is an educational platform focused on architecture and design, offering workshops and courses on advanced design tools, computational design, and AI integration in architecture. The platform provides insights, tutorials, and live events to inspire and educate aspiring designers. PAACADEMY features renowned instructors and covers topics such as parametric design, 3D printing, and AI-driven architectural practices.
EvolveLab
EvolveLab is a digital solutions provider specializing in BIM management and app development for the AEC (Architecture, Engineering, and Construction) industry. They offer a range of powerful apps and services designed to empower architects, engineers, and contractors to streamline their workflows and bring their ideas to life more efficiently. With a focus on data-driven design and AI technology, EvolveLab's innovative tools help users enhance productivity and turn concepts into reality.
Proscia
Proscia is a leading provider of digital pathology solutions for the modern laboratory. Its flagship product, Concentriq, is an enterprise pathology platform that enables anatomic pathology laboratories to achieve 100% digitization and deliver faster, more precise results. Proscia also offers a range of AI applications that can be used to automate tasks, improve diagnostic accuracy, and accelerate research. The company's mission is to perfect cancer diagnosis with intelligent software that changes the way the world practices pathology.
AIOZ Network
AIOZ Network is an AI-powered platform that focuses on Web3, AI, storage, and streaming services. It offers decentralized AI computation, fast and reliable storage solutions, and seamless video streaming for dApps within the network. AIOZ aims to empower a fast, secure, and decentralized future by providing a one-click integration of dApps on the AIOZ blockchain, supporting popular smart contract languages, and utilizing spare computing resources from a global community of nodes.
Live Portrait Ai Generator
Live Portrait Ai Generator is an AI application that transforms static portrait images into lifelike videos using advanced animation technology. Users can effortlessly animate their portraits, fine-tune animations, unleash artistic styles, and make memories move with text, music, and other elements. The tool offers a seamless stitching technology and retargeting capabilities to achieve perfect results. Live Portrait Ai enhances generation quality and generalization ability through a mixed image-video training strategy and network architecture upgrades.
Altera
Altera is an applied research company focused on building digital humans - machines with fundamental human qualities. Led by Dr. Robert Yang, the team comprises computational neuroscientists, CS and Physics experts from prestigious institutions. Their mission is to create digital human beings that can live, care, and grow with us. The company's early research prototypes began in games, offering a glimpse into the potential of these digital humans.
CCDS
CCDS (Center for Computational & Data Sciences) is a research center at Independent University Bangladesh dedicated to artificial intelligence, data sciences, and computational science. The center has various wings focusing on AI, computational biology, physics, data science, human-computer interaction, and industry partnerships. CCDS explores the use of computation to understand nature and society, uncover hidden stories in data, and tackle complex challenges. The center collaborates with institutions like CERN and the Dunlap Institute for Astronomy and Astrophysics.

Owkin
Owkin is a full-stack AI biotech company that integrates the best of human and artificial intelligence to deliver better drugs and diagnostics at scale. By understanding complex biology through AI, Owkin identifies new treatments, de-risks and accelerates clinical trials, and builds diagnostic tools to reduce time to impact for patients.

NLTK
NLTK (Natural Language Toolkit) is a leading platform for building Python programs to work with human language data. It provides easy-to-use interfaces to over 50 corpora and lexical resources such as WordNet, along with a suite of text processing libraries for classification, tokenization, stemming, tagging, parsing, and semantic reasoning, wrappers for industrial-strength NLP libraries, and an active discussion forum. Thanks to a hands-on guide introducing programming fundamentals alongside topics in computational linguistics, plus comprehensive API documentation, NLTK is suitable for linguists, engineers, students, educators, researchers, and industry users alike.
14 - Open Source Tools
matsciml
The Open MatSci ML Toolkit is a flexible framework for machine learning in materials science. It provides a unified interface to a variety of materials science datasets, as well as a set of tools for data preprocessing, model training, and evaluation. The toolkit is designed to be easy to use for both beginners and experienced researchers, and it can be used to train models for a wide range of tasks, including property prediction, materials discovery, and materials design.

NoLabs
NoLabs is an open-source biolab that provides easy access to state-of-the-art models for bio research. It supports various tasks, including drug discovery, protein analysis, and small molecule design. NoLabs aims to accelerate bio research by making inference models accessible to everyone.

AlphaFold3
AlphaFold3 is an implementation of the Alpha Fold 3 model in PyTorch for accurate structure prediction of biomolecular interactions. It includes modules for genetic diffusion and full model examples for forward pass computations. The tool allows users to generate random pair and single representations, operate on atomic coordinates, and perform structure predictions based on input tensors. The implementation also provides functionalities for training and evaluating the model.
crystal-text-llm
This repository contains the code for the paper Fine-Tuned Language Models Generate Stable Inorganic Materials as Text. It demonstrates how finetuned LLMs can be used to generate stable materials, match or exceed the performance of domain specific models, mutate existing materials, and sample crystal structures conditioned on text descriptions. The method is distinct from CrystaLLM, which trains language models from scratch on CIF-formatted crystals.

Scientific-LLM-Survey
Scientific Large Language Models (Sci-LLMs) is a repository that collects papers on scientific large language models, focusing on biology and chemistry domains. It includes textual, molecular, protein, and genomic languages, as well as multimodal language. The repository covers various large language models for tasks such as molecule property prediction, interaction prediction, protein sequence representation, protein sequence generation/design, DNA-protein interaction prediction, and RNA prediction. It also provides datasets and benchmarks for evaluating these models. The repository aims to facilitate research and development in the field of scientific language modeling.
md-agent
MD-Agent is a LLM-agent based toolset for Molecular Dynamics. It uses Langchain and a collection of tools to set up and execute molecular dynamics simulations, particularly in OpenMM. The tool assists in environment setup, installation, and usage by providing detailed steps. It also requires API keys for certain functionalities, such as OpenAI and paper-qa for literature searches. Contributions to the project are welcome, with a detailed Contributor's Guide available for interested individuals.
AIMNet2
AIMNet2 Calculator is a package that integrates the AIMNet2 neural network potential into simulation workflows, providing fast and reliable energy, force, and property calculations for molecules with diverse elements. It excels at modeling various systems, offers flexible interfaces for popular simulation packages, and supports long-range interactions using DSF or Ewald summation Coulomb models. The tool is designed for accurate and versatile molecular simulations, suitable for large molecules and periodic calculations.
admet_ai
ADMET-AI is a platform for ADMET prediction using Chemprop-RDKit models trained on ADMET datasets from the Therapeutics Data Commons. It offers command line, Python API, and web server interfaces for making ADMET predictions on new molecules. The platform can be easily installed using pip and supports GPU acceleration. It also provides options for processing TDC data, plotting results, and hosting a web server. ADMET-AI is a machine learning platform for evaluating large-scale chemical libraries.
awesome-AI4MolConformation-MD
The 'awesome-AI4MolConformation-MD' repository focuses on protein conformations and molecular dynamics using generative artificial intelligence and deep learning. It provides resources, reviews, datasets, packages, and tools related to AI-driven molecular dynamics simulations. The repository covers a wide range of topics such as neural networks potentials, force fields, AI engines/frameworks, trajectory analysis, visualization tools, and various AI-based models for protein conformational sampling. It serves as a comprehensive guide for researchers and practitioners interested in leveraging AI for studying molecular structures and dynamics.
AI2BMD
AI2BMD is a program for efficiently simulating protein molecular dynamics with ab initio accuracy. The repository contains datasets, simulation programs, and public materials related to AI2BMD. It provides a Docker image for easy deployment and a standalone launcher program. Users can run simulations by downloading the launcher script and specifying simulation parameters. The repository also includes ready-to-use protein structures for testing. AI2BMD is designed for x86-64 GNU/Linux systems with recommended hardware specifications. The related research includes model architectures like ViSNet, Geoformer, and fine-grained force metrics for MLFF. Citation information and contact details for the AI2BMD Team are provided.
AIRS
AIRS is a collection of open-source software tools, datasets, and benchmarks focused on Artificial Intelligence for Science in Quantum, Atomistic, and Continuum Systems. The goal is to develop and maintain an integrated, open, reproducible, and sustainable set of resources to advance the field of AI for Science. The current resources include tools for Quantum Mechanics, Density Functional Theory, Small Molecules, Protein Science, Materials Science, Molecular Interactions, and Partial Differential Equations.

chem-bench
ChemBench is a project aimed at expanding chemistry benchmark tasks in a BIG-bench compatible way, providing a pipeline to benchmark frontier and open models. It allows users to run benchmarking tasks on models with existing presets, offering predefined parameters and processing steps. The library facilitates benchmarking models on the entire suite, addressing challenges such as prompt structure, parsing, and scoring methods. Users can contribute to the project by following the developer notes.
gromacs_copilot
GROMACS Copilot is an agent designed to automate molecular dynamics simulations for proteins in water using GROMACS. It handles system setup, simulation execution, and result analysis automatically, providing outputs such as RMSD, RMSF, Rg, and H-bonds. Users can interact with the agent through prompts and API keys from DeepSeek and OpenAI. The tool aims to simplify the process of running MD simulations, allowing users to focus on other tasks while it handles the technical aspects of the simulations.
ai2-kit
A toolkit for computational chemistry research, featuring tools to facilitate automated workflows. Includes tools for NMR prediction, dynamic catalysis research, proton transfer analysis, amorphous oxides structure analysis, reweighting, and more. Users can install 'ai2-kit' via pip and explore various domain-specific and general tools for processing system data and filtering structures by model deviation.
16 - OpenAI Gpts
Formula Generator
Expert in generating and explaining mathematical, chemical, and computational formulas.

StephenBot
A digital homage to honor Stephen Wolfram's impact on computational science and technology and to celebrate his dedication to public education, powered by Stephen Wolfram's wealth of public presentations, writings, and live streams.
ChatPNP
Blends academic insights & accessible explanations on P vs NP, drawing from Lance Fortnow's works.