
nmed2024
AI-based differential diagnosis of dementia etiologies on multimodal data
Stars: 73

Nmed2024 is a GitHub repository that contains code for a neural network model designed for medical image analysis. The repository includes scripts for training the model, as well as pre-trained weights for quick deployment. The model is specifically tailored for detecting abnormalities in medical images, such as tumors or fractures. It utilizes deep learning techniques to achieve high accuracy and can be easily integrated into existing medical imaging systems. Researchers and developers in the healthcare industry can leverage this tool to enhance the efficiency and accuracy of medical image analysis tasks.
README:
This work is published in Nature Medicine (https://doi.org/10.1038/s41591-024-03118-z).
This repository contains the implementation of a deep learning framework for the differential diagnosis of dementia etiologies using multi-modal data. Using data from $9$ distinct cohorts totalling $51,269$ participants, we developed an algorithmic framework, utilizing transformers and self-supervised learning, to execute differential diagnoses of dementia. This model classifies individuals into one or more of thirteen meticulously curated diagnostic categories, each aligning closely with real-world clinical requirements. These categories span the entire spectrum of cognitive conditions, from normal cognition (NC), mild cognitive impairment (MCI) to dementia (DE), and further include $10$ dementia types.
Figure 1: Data, model architecture, and modeling strategy. (a) Our model for differential dementia diagnosis was developed using diverse data modalities, including individual-level demographics, health history, neurological testing, physical/neurological exams, and multi-sequence MRI scans. These data sources whenever available were aggregated from nine independent cohorts: 4RTNI, ADNI, AIBL, FHS, LBDSU, NACC, NIFD, OASIS, and PPMI. For model training, we merged data from NACC, AIBL, PPMI, NIFD, LBDSU, OASIS and 4RTNI. We employed a subset of the NACC dataset for internal testing. For external validation, we utilized the ADNI and FHS cohorts. (b) A transformer served as the scaffold for the model. Each feature was processed into a fixed-length vector using a modality-specific embedding strategy and fed into the transformer as input. A linear layer was used to connect the transformer with the output prediction layer. (c) A distinct portion of the NACC dataset was randomly selected to enable a comparative analysis of the model’s performance against practicing neurologists. Furthermore, we conducted a direct comparison between the model and a team of practicing neuroradiologists using a random sample of cases with confirmed dementia from the NACC testing cohort. For both these evaluations, the model and clinicians had access to the same set of multimodal data. Finally, we assessed the model’s predictions by comparing them with pathology grades available from the NACC, ADNI, and FHS cohorts.
To setup the adrd package, run the following in the root of the repository:
pip install git+https://github.com/vkola-lab/nmed2024.gitThe tool was developed using the following dependencies:
- Python (3.11.7 or greater)
- PyTorch (2.1 or greater).
- TorchIO (0.15 or greater).
- MONAI (1.1 or greater).
- NumPy (1.24 or greater).
- tqdm (4.62 or greater).
- pandas (1.5.3 or greater).
- nibabel (5.0 or greater).
- matplotlib (3.7.2 or greater).
- shap (0.43 or greater).
- scikit-learn (1.2.2 or greater).
- scipy (1.10 or greater).
You can clone this repository using the following command:
git clone https://github.com/vkola-lab/nmed2024.gitThe training process consists of two stages:
All code related to training the imaging model with self-supervised learning is under ./dev/ssl_mri/.
Note: we used skull stripped MRIs to get our image embeddings. We have provided the script for skull stripping using the publicly available SynthStrip tool [2]. The code is provided under dev/skullstrip.sh.
We trained started from the self-supervised pre-trained weights of the Swin UNETR encoder (CVPR paper [1]) which can be downloaded from this link. The checkpoint should be saved under ./dev/ssl_mri/pretrained_models/.
To finetune the pre-trained Swin UNETR on your own data, run the following commands:
cd dev/ssl_mri/
bash scripts/run_swinunetr.shThe code can run in a multi-GPU setting by setting --nproc_per_node to the appropriate number of available GPUs.
Once a finetuned checkpoint of the imaging model is saved, navigate to the repository's root directory and run dev/train.sh with the following changes in flag values:
img_net="SwinUNETR"
img_mode=2 # loads the imgnet, generates embeddings out of the MRIs input to the network, and saves them.
Once image embeddings are saved, we train the backbone transformer on the multi-modal data. Create a configuration file similar to default_conf_new.toml, categorizing each feature as numerical, categorical or imaging. Please add the saved image embedding paths to your data file as another column and set the type of this feature as imaging in the configuration file.
Navigate to the repository's root directory and run dev/train.sh with the following changes in flag values:
img_net="SwinUNETREMB"
img_mode=1 # loads MRI embeddings and not the imgnet.To train the model without imaging, please use the following flag values:
img_net="NonImg"
img_mode=-1The model predictions were generated using the script dev/generate_predictions.py. All AUC-ROC curves and AUC-PR curves were generated using plots/roc_pr_curves.py.
To make our deep learning framework for differential dementia diagnosis more accessible and user-friendly, we have hosted it on Huggingface Space. This interactive demo allows users to experience the power and efficiency of our model in real-time, providing an intuitive interface for uploading diagnostic information and receiving diagnostic predictions. Check out our Huggingface demo https://huggingface.co/spaces/vkola-lab/nmed2024 to see our model in action and explore its potential.
[1] Tang, Y., Yang, D., Li, W., Roth, H.R., Landman, B., Xu, D., Nath, V. and Hatamizadeh, A., 2022. Self-supervised pre-training of swin transformers for 3d medical image analysis. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (pp. 20730-20740).
[2] Hoopes, A., Mora, J.S., Dalca, A.V., Fischl, B. and Hoffmann, M., 2022. SynthStrip: Skull-stripping for any brain image. NeuroImage, 260, p.119474.
@article{xue2024ai,
title={AI-based differential diagnosis of dementia etiologies on multimodal data},
author={Xue, Chonghua and Kowshik, Sahana S and Lteif, Diala and Puducheri, Shreyas and Jasodanand, Varuna H and Zhou, Olivia T and Walia, Anika S and Guney, Osman B and Zhang, J Diana and Pham, Serena T and others},
journal={Nature Medicine},
pages={1--13},
year={2024},
publisher={Nature Publishing Group US New York}
}
For Tasks:
Click tags to check more tools for each tasksFor Jobs:
Alternative AI tools for nmed2024
Similar Open Source Tools

nmed2024
Nmed2024 is a GitHub repository that contains code for a neural network model designed for medical image analysis. The repository includes scripts for training the model, as well as pre-trained weights for quick deployment. The model is specifically tailored for detecting abnormalities in medical images, such as tumors or fractures. It utilizes deep learning techniques to achieve high accuracy and can be easily integrated into existing medical imaging systems. Researchers and developers in the healthcare industry can leverage this tool to enhance the efficiency and accuracy of medical image analysis tasks.
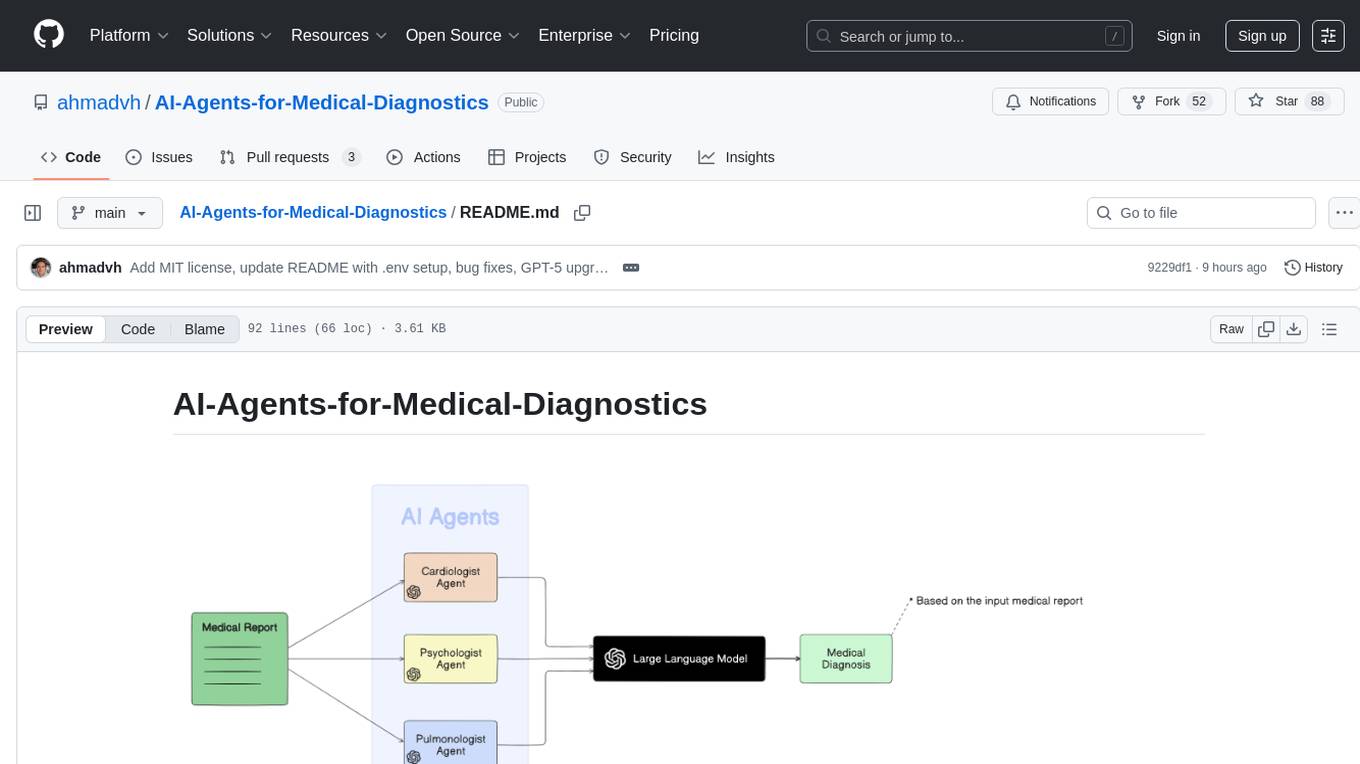
AI-Agents-for-Medical-Diagnostics
AI Agents for Medical Diagnostics is a repository containing a collection of machine learning models and algorithms designed to assist in medical diagnosis. The tools provided in this repository are specifically tailored for analyzing medical data and making predictions related to various health conditions. By leveraging the power of artificial intelligence, these agents aim to improve the accuracy and efficiency of diagnostic processes in the medical field. Researchers, healthcare professionals, and data scientists can benefit from the resources available in this repository to develop innovative solutions for diagnosing illnesses and predicting patient outcomes.
grand-challenge.org
Grand Challenge is a platform that provides access to large amounts of annotated training data, objective comparisons of state-of-the-art machine learning solutions, and clinical validation using real-world data. It assists researchers, data scientists, and clinicians in collaborating to develop robust machine learning solutions to problems in biomedical imaging.
AI_Spectrum
AI_Spectrum is a versatile machine learning library that provides a wide range of tools and algorithms for building and deploying AI models. It offers a user-friendly interface for data preprocessing, model training, and evaluation. With AI_Spectrum, users can easily experiment with different machine learning techniques and optimize their models for various tasks. The library is designed to be flexible and scalable, making it suitable for both beginners and experienced data scientists.

deeppowers
Deeppowers is a powerful Python library for deep learning applications. It provides a wide range of tools and utilities to simplify the process of building and training deep neural networks. With Deeppowers, users can easily create complex neural network architectures, perform efficient training and optimization, and deploy models for various tasks. The library is designed to be user-friendly and flexible, making it suitable for both beginners and experienced deep learning practitioners.
Awesome-Efficient-MoE
Awesome Efficient MoE is a GitHub repository that provides an implementation of Mixture of Experts (MoE) models for efficient deep learning. The repository includes code for training and using MoE models, which are neural network architectures that combine multiple expert networks to improve performance on complex tasks. MoE models are particularly useful for handling diverse data distributions and capturing complex patterns in data. The implementation in this repository is designed to be efficient and scalable, making it suitable for training large-scale MoE models on modern hardware. The code is well-documented and easy to use, making it accessible for researchers and practitioners interested in leveraging MoE models for their deep learning projects.
lemonai
LemonAI is a versatile machine learning library designed to simplify the process of building and deploying AI models. It provides a wide range of tools and algorithms for data preprocessing, model training, and evaluation. With LemonAI, users can easily experiment with different machine learning techniques and optimize their models for various tasks. The library is well-documented and beginner-friendly, making it suitable for both novice and experienced data scientists. LemonAI aims to streamline the development of AI applications and empower users to create innovative solutions using state-of-the-art machine learning methods.
deepteam
Deepteam is a powerful open-source tool designed for deep learning projects. It provides a user-friendly interface for training, testing, and deploying deep neural networks. With Deepteam, users can easily create and manage complex models, visualize training progress, and optimize hyperparameters. The tool supports various deep learning frameworks and allows seamless integration with popular libraries like TensorFlow and PyTorch. Whether you are a beginner or an experienced deep learning practitioner, Deepteam simplifies the development process and accelerates model deployment.

pdr_ai_v2
pdr_ai_v2 is a Python library for implementing machine learning algorithms and models. It provides a wide range of tools and functionalities for data preprocessing, model training, evaluation, and deployment. The library is designed to be user-friendly and efficient, making it suitable for both beginners and experienced data scientists. With pdr_ai_v2, users can easily build and deploy machine learning models for various applications, such as classification, regression, clustering, and more.
minions
Minions is a communication protocol that enables small on-device models to collaborate with frontier models in the cloud. By only reading long contexts locally, it reduces cloud costs with minimal or no quality degradation. The repository provides a demonstration of the protocol.
dranet
Dranet is a Python library for analyzing and visualizing data from neural networks. It provides tools for interpreting model predictions, understanding feature importance, and evaluating model performance. With Dranet, users can gain insights into how neural networks make decisions and improve model transparency and interpretability.
sciml.ai
SciML.ai is an open source software organization dedicated to unifying packages for scientific machine learning. It focuses on developing modular scientific simulation support software, including differential equation solvers, inverse problems methodologies, and automated model discovery. The organization aims to provide a diverse set of tools with a common interface, creating a modular, easily-extendable, and highly performant ecosystem for scientific simulations. The website serves as a platform to showcase SciML organization's packages and share news within the ecosystem. Pull requests are encouraged for contributions.
God-Level-AI
A drill of scientific methods, processes, algorithms, and systems to build stories & models. An in-depth learning resource for humans. This repository is designed for individuals aiming to excel in the field of Data and AI, providing video sessions and text content for learning. It caters to those in leadership positions, professionals, and students, emphasizing the need for dedicated effort to achieve excellence in the tech field. The content covers various topics with a focus on practical application.
FLAME
FLAME is a lightweight and efficient deep learning framework designed for edge devices. It provides a simple and user-friendly interface for developing and deploying deep learning models on resource-constrained devices. With FLAME, users can easily build and optimize neural networks for tasks such as image classification, object detection, and natural language processing. The framework supports various neural network architectures and optimization techniques, making it suitable for a wide range of applications in the field of edge computing.
ml-retreat
ML-Retreat is a comprehensive machine learning library designed to simplify and streamline the process of building and deploying machine learning models. It provides a wide range of tools and utilities for data preprocessing, model training, evaluation, and deployment. With ML-Retreat, users can easily experiment with different algorithms, hyperparameters, and feature engineering techniques to optimize their models. The library is built with a focus on scalability, performance, and ease of use, making it suitable for both beginners and experienced machine learning practitioners.
For similar tasks

nmed2024
Nmed2024 is a GitHub repository that contains code for a neural network model designed for medical image analysis. The repository includes scripts for training the model, as well as pre-trained weights for quick deployment. The model is specifically tailored for detecting abnormalities in medical images, such as tumors or fractures. It utilizes deep learning techniques to achieve high accuracy and can be easily integrated into existing medical imaging systems. Researchers and developers in the healthcare industry can leverage this tool to enhance the efficiency and accuracy of medical image analysis tasks.
Caissa
Caissa is a strong, UCI command-line chess engine optimized for regular chess, FRC, and DFRC. It features its own neural network trained with self-play games, supports various UCI options, and provides different EXE versions for different CPU architectures. The engine uses advanced search algorithms, neural network evaluation, and endgame tablebases. It offers outstanding performance in ultra-short games and is written in C++ with modules for backend, frontend, and utilities like neural network trainer and self-play data generator.
BetaML.jl
The Beta Machine Learning Toolkit is a package containing various algorithms and utilities for implementing machine learning workflows in multiple languages, including Julia, Python, and R. It offers a range of supervised and unsupervised models, data transformers, and assessment tools. The models are implemented entirely in Julia and are not wrappers for third-party models. Users can easily contribute new models or request implementations. The focus is on user-friendliness rather than computational efficiency, making it suitable for educational and research purposes.
For similar jobs
Taiyi-LLM
Taiyi (太一) is a bilingual large language model fine-tuned for diverse biomedical tasks. It aims to facilitate communication between healthcare professionals and patients, provide medical information, and assist in diagnosis, biomedical knowledge discovery, drug development, and personalized healthcare solutions. The model is based on the Qwen-7B-base model and has been fine-tuned using rich bilingual instruction data. It covers tasks such as question answering, biomedical dialogue, medical report generation, biomedical information extraction, machine translation, title generation, text classification, and text semantic similarity. The project also provides standardized data formats, model training details, model inference guidelines, and overall performance metrics across various BioNLP tasks.

nmed2024
Nmed2024 is a GitHub repository that contains code for a neural network model designed for medical image analysis. The repository includes scripts for training the model, as well as pre-trained weights for quick deployment. The model is specifically tailored for detecting abnormalities in medical images, such as tumors or fractures. It utilizes deep learning techniques to achieve high accuracy and can be easily integrated into existing medical imaging systems. Researchers and developers in the healthcare industry can leverage this tool to enhance the efficiency and accuracy of medical image analysis tasks.
fuse-med-ml
FuseMedML is a Python framework designed to accelerate machine learning-based discovery in the medical field by promoting code reuse. It provides a flexible design concept where data is stored in a nested dictionary, allowing easy handling of multi-modality information. The framework includes components for creating custom models, loss functions, metrics, and data processing operators. Additionally, FuseMedML offers 'batteries included' key components such as fuse.data for data processing, fuse.eval for model evaluation, and fuse.dl for reusable deep learning components. It supports PyTorch and PyTorch Lightning libraries and encourages the creation of domain extensions for specific medical domains.
MedLLMsPracticalGuide
This repository serves as a practical guide for Medical Large Language Models (Medical LLMs) and provides resources, surveys, and tools for building, fine-tuning, and utilizing LLMs in the medical domain. It covers a wide range of topics including pre-training, fine-tuning, downstream biomedical tasks, clinical applications, challenges, future directions, and more. The repository aims to provide insights into the opportunities and challenges of LLMs in medicine and serve as a practical resource for constructing effective medical LLMs.
hi-ml
The Microsoft Health Intelligence Machine Learning Toolbox is a repository that provides low-level and high-level building blocks for Machine Learning / AI researchers and practitioners. It simplifies and streamlines work on deep learning models for healthcare and life sciences by offering tested components such as data loaders, pre-processing tools, deep learning models, and cloud integration utilities. The repository includes two Python packages, 'hi-ml-azure' for helper functions in AzureML, 'hi-ml' for ML components, and 'hi-ml-cpath' for models and workflows related to histopathology images.
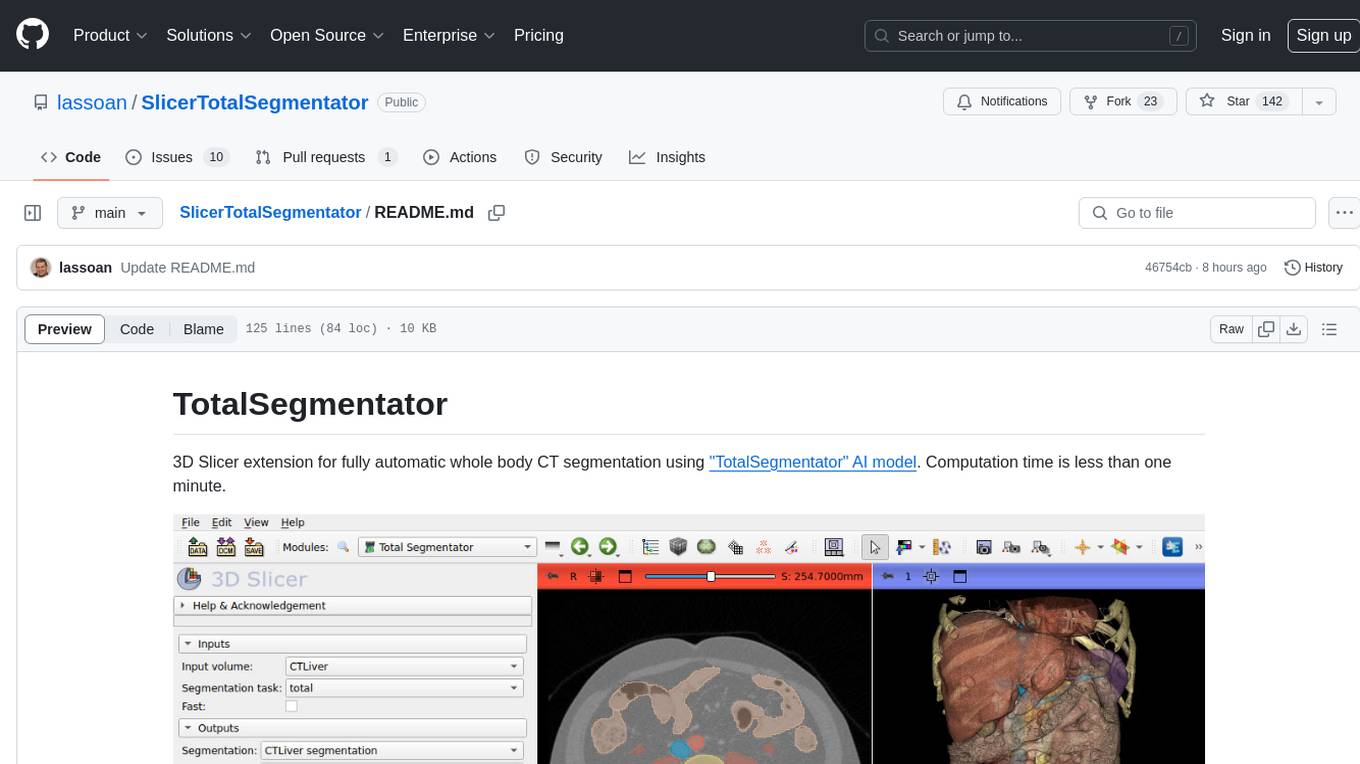
SlicerTotalSegmentator
TotalSegmentator is a 3D Slicer extension designed for fully automatic whole body CT segmentation using the 'TotalSegmentator' AI model. The computation time is less than one minute, making it efficient for research purposes. Users can set up GPU acceleration for faster segmentation. The tool provides a user-friendly interface for loading CT images, creating segmentations, and displaying results in 3D. Troubleshooting steps are available for common issues such as failed computation, GPU errors, and inaccurate segmentations. Contributions to the extension are welcome, following 3D Slicer contribution guidelines.
machine-learning-research
The 'machine-learning-research' repository is a comprehensive collection of resources related to mathematics, machine learning, deep learning, artificial intelligence, data science, and various scientific fields. It includes materials such as courses, tutorials, books, podcasts, communities, online courses, papers, and dissertations. The repository covers topics ranging from fundamental math skills to advanced machine learning concepts, with a focus on applications in healthcare, genetics, computational biology, precision health, and AI in science. It serves as a valuable resource for individuals interested in learning and researching in the fields of machine learning and related disciplines.
LLMonFHIR
LLMonFHIR is an iOS application that utilizes large language models (LLMs) to interpret and provide context around patient data in the Fast Healthcare Interoperability Resources (FHIR) format. It connects to the OpenAI GPT API to analyze FHIR resources, supports multiple languages, and allows users to interact with their health data stored in the Apple Health app. The app aims to simplify complex health records, provide insights, and facilitate deeper understanding through a conversational interface. However, it is an experimental app for informational purposes only and should not be used as a substitute for professional medical advice. Users are advised to verify information provided by AI models and consult healthcare professionals for personalized advice.


