AI-Drug-Discovery-Design
AI drug design
Stars: 77
AI-Drug-Discovery-Design is a repository focused on Artificial Intelligence-assisted Drug Discovery and Design. It explores the use of AI technology to accelerate and optimize the drug development process. The advantages of AI in drug design include speeding up research cycles, improving accuracy through data-driven models, reducing costs by minimizing experimental redundancies, and enabling personalized drug design for specific patients or disease characteristics.
README:
让我为大家简要介绍一下我负责的领域——人工智能辅助药物发现与设计。为了更好地帮助大家理解这一复杂的前沿技术,我将通过中文为大家进行讲解,特别是为那些可能在语言上有所障碍的同学提供支持。尽管这种内容通常更适合以英语呈现,但为了确保大家能够更好地掌握核心概念并迅速上手,我选择用中文为大家提供更加直观的学习路径。
人工智能辅助药物发现与设计是一种利用人工智能(AI)技术来加速和优化药物开发过程的方法。传统药物研发周期长、成本高、成功率低,而 AI 能够通过大数据处理、模型预测和自动化分析,大幅提升药物设计的效率和准确性。
- 加速研发周期:AI 能够快速筛选大量化合物,减少实验筛选的工作量。
- 提高准确性:AI 能够通过大规模数据训练模型,提升药物设计中的预测准确性。
- 成本降低:由于减少了实验的重复性和失败率,AI 能够有效降低药物研发成本。
- 个性化药物设计:AI 能帮助设计个性化药物,针对特定患者或疾病特征,优化治疗效果。
人工智能在药物发现与设计中的应用,使得药物开发过程更加智能化、自动化。通过化合物筛选、靶标预测、药物生成、ADMET 分析等关键步骤,AI 能有效加速药物研发的进程,同时提高设计的准确性和效率。
- 首先我感谢我的中山大学医学院的导师雷教授带我入门这一行,我踏入校园一无所知,所以也不知道自己的方向,一脸茫然的,所以还是特别感谢我的导师的。并且还要感谢淮阴工学院——喻教授,在他的帮助下才能写出来。同时也感谢张胜玉同学的大力支持完成学业。
- 第二个首先感谢,南京工业大学的计算化学大佬——“郭为涛”同学,一直帮助我,给我讲解制药的流程。
- 第三,我还是得感谢我的好朋友——南京医科大学的“周仪萍”同学,是她聪明漂亮善良的姑娘,大力帮助我。
- 其次,感谢我的群里面的大佬各路指导。
- 最后感谢社会人士和同道中人来帮我批评指正,让我的这一份文档,这一份代码写得更完整,更加完善,让我们祖国做的更好更强大,感谢祖国,感谢党。
- 感谢这些帮助我的人完成我的学业!
Abstract: Drug discovery and development affects various aspects of human health and dramatically impacts the pharmaceutical market. However, investments in a new drug often go unrewarded due to the long and complex process of drug research and development (R&D). With the advancement of experimental technology and computer hardware, artificial intelligence (AI) has recently emerged as a leading tool in analyzing abundant and high-dimensional data. Explosive growth in the size of biomedical data provides advantages in applying AI in all stages of drug R&D. Driven by big data in biomedicine, AI has led to a revolution in drug R&D, due to its ability to discover new drugs more efficiently and at lower cost. This review begins with a brief overview of common AI models in the field of drug discovery; then, it summarizes and discusses in depth their specific applications in various stages of drug R&D, such as target discovery, drug discovery and design, preclinical research, automated drug synthesis, and influences in the pharmaceutical market. Finally, the major limitations of AI in drug R&D are fully discussed and possible solutions are proposed.
Keywords: Artificial intelligence; Machine learning; Deep learning; Target identification; Target discovery; Drug design; Drug discovery
- 👉🏻👉🏻👉🏻👉🏻👉🏻目前目录分为了,第一、二节和第三节分别:提供没有基础的同学,有基础的同学
-
人工智能在医学中的应用
- Python基础
- Numpy、Pandas
- Matplotlib
- 机器学习和Scikit-Learn
- 深度学习
- CADD
- 图神经网络
-
Python和机器学习基础
- 分子的文本表示:SMILES
- 分子的向量表示:描述符和指纹
- RDKit简介
- 经典机器学习模型:线性回归、随机森林、支持向量机
-
公开可用的小分子数据集的探索
- 生物活性分子 ChEMBL 数据库
- ZINC 数据库
- PubChem 数据库
- 探索性数据分析 (EDA)
- 定量构效关系 (QSAR) 和虚拟筛选 (VS)
-
图神经网络
- 神经网络架构和训练
- 分子图、原子特征化
- 消息传递神经网络
- 图卷积神经网络
- 可解释性:Grad-CAM
-
分子对接
- 分子数据格式:SMI、SDF、MOL2、PDB
- 力场
- 蛋白质折叠
- 使用 AutoDock Vina、Smina、QuickVina 进行分子对接
- 交互指纹
- 药效团
-
深度生成模型
- 自动编码器
- 循环神经网络
- SMILES生成器:ReLeaSE 和 REINVENT
- 基于图的生成模型:JT-VAE
- 分子特性优化:强化学习和贝叶斯优化
-
蛋白质深度学习
- 简化的蛋白质图表示
- 体素网格表示
- 用于编码蛋白质表面的网格表示
- 3D卷积神经网络
-
不确定性预测
- 任意和认知的不确定性
- 共形预测
-
基础知识
- 化学信息学 RDKit 简介
- Pandas 在化学信息学中的应用
- SMILES 教程
- SMARTS 教程
- 反应列举基础知识
- 立体异构体和互变异构体列举
-
使用 Datamol 和 Molfeat 精简化学信息学工作流
- 数据处理、描述符和聚类
-
聚类
- K-Means 聚类
- Taylor-Butina 聚类
-
复杂的化学信息学分析
- Chembl 系统分析
- 基于 Chembl 数据库的药物数据分析
- 基于 BindingDB 中的专利数据进行分析
-
SAR 分析
- 脚手架识别
- R-group 分析
- 位置模拟扫描分析
- Free-Wilson 分析
- 匹配的分子对
- 匹配的分子集
-
机器学习模型
- 构建并测试一个 QSAR 模型
- 分类模型构建与比较
- 回归模型构建与比较
-
主动学习
- 主动分类
- 主动回归
- 主动形状搜索
-
神经网络潜能
- 使用 Auto3D 的同分异构体能量预测
- 01_从 ChEMBL 化合物数据采集
- 02_从 PubChem 获取数据
- 03_从 KLIFS 获取数据
- 03_1 完整项目:《基于机器学习的生物活性预测》
- 04_查询在线 API 网络服务
- 05_分子过滤:ADMET 和先导化合物相似标准
- 05_1 完整项目:《基于机器学习与深度学习的分子ADMET预测》
- 05_2 完整项目:《基于GNN的分子毒性预测》
- 06_分子过滤:不需要的子结构
- 06_1 完整项目:《基于ADMET和RO5的分子筛选与化合物相似性的配体筛选》
- 07_分子表示
- 08_基于配体的筛选:化合物相似性
- 09_复合聚类
- 10_最大公共子结构
- 11_基于配体的药效团
- 12_结合位点相似性和脱靶预测
- 13_蛋白质数据获取:蛋白质数据库(PDB)
- 14_结合位点检测
- 15_蛋白质-配体对接
- 15_1 预测生物活性分子的逆合成可及性
- 16_蛋白质-配体相互作用
- 17_NGLview 高级使用
- 18_分子动力学模拟
- 19_分析分子动力学模拟
- 20_先导化合物优化的自动化流程
- 21_基于配体的筛选:机器学习
- 22_基于配体的筛选:神经网络
- 23_基于 RNN 的分子性质预测
- 24_基于 GNN 的分子性质预测
- 25_分子特性预测转换器
- 26_不确定性估计
-
27_1 RNA Aptamer 数据来源
- 数据来源:RNAapt3D (https://rnaapt3d.medals.jp/)
- 27_2 数据清洗与预处理
-
28_1 一级结构预测
-
28_2 结构可视化与分析
-
29_1 二级结构预测
-
29_2 能量最小化与折叠稳定性分析
- ΔG(自由能)和折叠稳定性图
- 30_1 RNA Aptamer 与靶标的结合位点预测
- 30_2 结合能计算与优化
- 30_3 基于 RNA Aptamer 的药物设计
- 30_4 药物化学与虚拟筛选
- 30_5 分子动力学模拟
- 30_6 实验验证
- 31_激酶相似性:序列
- 32_激酶相似性:激酶口袋(KiSSim 指纹)
- 33_激酶相似性:相互作用指纹
- 34_激酶相似性:配体概况
- 35_激酶相似性:不同观点比较
- 36_基于激酶片段库设计激酶抑制剂
- 37_蛋白质-配体相互作用预测
- 完整项目:《项目实战:基于Transformer的有机化学反应产量预测 (Prediction of chemical reaction yields using deep learning)》
- 完整项目:《项目实战:Mapping the space of chemical reactions using attention-based neural networks》
- 完整项目:《项目实战:基于图数据的小分子化合物生成模型(A Graph to Graphs Framework for Retrosynthesis Prediction)》
- 完整项目:《项目实战:基于NLP的抗体生成模型(Generative language modeling for antibody design)》
- 38_基于 KLIFS 数据跑 3D 动力学
- 39_基于共识对接的一体化结构虚拟筛选(蛋白质制备、对接、结合位点选择、重新评分和排序)
- 40_One-Hot 编码
- 41_使用代码绘制分子图
- 研究目标:探索基因与免疫浸润之间的关系
- 背景介绍:免疫浸润的重要性及其在基因表达中的影响
- 安装与配置所需的软件工具(如 R、Python、Bioconductor )
- 从 GEO、TCGA 等数据库下载基因表达数据
- 数据标准化和清洗
- 使用注释文件(如 GTF 或 GFF 文件)对基因表达数据进行注释
- 工具:
biomaRt、org.Hs.eg.db
- 使用 SVA 校正批次效应
- 代码实现:
sva包中的ComBat或svaseq方法
- 使用
limma、DESeq2等工具进行差异基因分析 - 生成差异表达基因(DEGs)列表
- 使用差异分析的结果绘制火山图
- 代码实现:
ggplot2、EnhancedVolcano包
-
08. Metascape
- 在 Metascape 网站上进行通路和功能富集分析
-
09. Gene Ontology (GO)
- 使用 GO 进行基因功能注释和富集分析
- 工具:
clusterProfiler、topGO
-
10. KEGG 富集分析
- 使用 KEGG 数据库进行信号通路富集分析
- 工具:
clusterProfiler、KEGGREST
-
11. Protein-Protein Interaction (PPI)
- 构建蛋白质-蛋白质互作网络
- 工具:
STRING数据库、Cytoscape
- 构建随机森林模型,筛选重要基因
- 绘制差异基因的表达热图
- 根据差异基因计算基因评分
- 代码实现:基于基因表达值的综合评分方法
- 使用神经网络模型对基因表达进行预测
- 工具:
neuralnet包
- 绘制 ROC 曲线,评估模型的准确性
- 工具:
pROC包
- 测试基因评分模型的准确性和可行性
- 测试神经网络模型的预测能力
- 使用测试数据集验证 ROC 曲线
- 使用 CIBERSORT 分析基因表达数据,估算免疫细胞的比例
- 绘制免疫细胞比例的柱状图
- 工具:
ggplot2
- 绘制基因表达与免疫细胞浸润之间的关系图
|-- Al-drug-design-reference.Data <- 参考文献文件夹
|-- README.md <- 详细简介
|-- img <- md的图片
|-- docs <- 文档
|-- Al-drug-design-reference.enl <- 参考文献文件
|-- list <- 项目结构目录
|-- Al-drug-design.yml <- 环境配置
| |-- 00_ai in_medicine <- Python基础知识(❤️如果你有Python基础,或者你有Python与药物设计基础,你可以跳过这一章节,直接从01开始看)
| |-- 01_Compound_data_acquisition <- 化合物采集
--------------------------------------
应安装 Anaconda 和 Git。请参阅Anaconda 的网站和Git 的网站进行下载。
❤️ u must need read paper
Where r u AIDrugDesign.yml ?
u first git clone my link!!!, it is have AIDrugDesign.yml.
conda env create -f AIDrugDesign.yml如果你在国外或者有🪜,请您打开 AIDrugDesign.yml
删除:
- pytorch
- https://mirrors.tuna.tsinghua.edu.cn/anaconda/pkgs/main
- https://mirrors.tuna.tsinghua.edu.cn/anaconda/pkgs/freeconda env listCheck to see if the AI-drug-design
conda activate AIDrugDesign------------------------------------------
如果你想要了解 更多的Git的知识,请您前往[如果操作git流程.md]
接下来,我将一步步为你讲解如何在 Windows 和 macOS 系统上安装和配置 Git,并拉取我的代码到本地。
- 前往 Git 官方下载页面。
- 选择 Windows 版本进行下载,并按照提示完成安装。
- 安装时,默认选项即可。如果想要自定义,可以根据需要选择不同的配置,比如编辑器、环境变量等。
安装完成后,打开终端(或 Git Bash)并配置你的 Git 用户信息:
git config --global user.name "Your Name"
git config --global user.email "[email protected]"这样,Git 就会在你每次提交代码时使用这些信息来标识你的身份。
在 Windows 上,打开 Git Bash,输入以下命令生成 SSH 密钥:
ssh-keygen -t rsa -b 4096 -C "[email protected]"- 按回车后,你会看到提示选择存储密钥的位置,默认按回车即可。
- 然后你需要设置一个密码,可以为空,但建议设置。
- 使用以下命令显示你生成的公钥:
cat ~/.ssh/id_rsa.pub- 复制公钥并登录 GitHub。
- 前往 GitHub 的 SSH 和 GPG 密钥页面,点击 New SSH key。
- 将你刚才复制的公钥粘贴到文本框中,添加后保存。
完成 SSH 密钥配置后,你可以使用以下命令克隆代码仓库:
git clone [email protected]:itWangCode/AI-drug-design.git运行命令后,Git 会将代码拉取到本地的文件夹中。如果成功,你将看到类似如下的信息:
Cloning into 'AI-drug-design'...这时,代码已经拉取成功。
macOS 上通常已经自带 Git,如果没有,可以通过 Homebrew 安装 Git:
- 打开终端,输入以下命令安装 Homebrew(如果没有安装):
/bin/bash -c "$(curl -fsSL https://raw.githubusercontent.com/Homebrew/install/HEAD/install.sh)"- 安装完成后,输入以下命令安装 Git:
brew install git- 打开终端,输入以下命令配置 Git:
git config --global user.name "Your Name"
git config --global user.email "[email protected]"- 在终端中输入以下命令生成 SSH 密钥:
ssh-keygen -t rsa -b 4096 -C "[email protected]"- 跟 Windows 一样,按回车使用默认路径存储密钥,设置密码(可选)。
- 在终端中使用以下命令查看公钥:
cat ~/.ssh/id_rsa.pub- 复制输出的公钥,登录 GitHub,将公钥添加到 GitHub SSH Keys 页面。
使用以下命令克隆你的仓库到本地:
git clone [email protected]:itWangCode/AI-drug-design.gitGit 会将代码下载到当前目录,表示代码拉取成功。
这样,你就成功配置了 Git 并克隆了代码仓库。
- 前往 Anaconda 官方网站。
- 点击下载按钮,选择适合 Windows 系统的版本(通常为 64-bit)。
- 下载完成后,运行安装程序并按照提示完成安装。
- 同样前往 Anaconda 官方网站。
- 选择 macOS 系统的版本下载并安装。
安装过程中,建议勾选 "Add Anaconda to my PATH environment variable" 选项,以便在终端中可以直接使用 conda 命令。
安装完成后:
- 在 Windows 上,可以通过开始菜单找到 Anaconda-Navigator 并运行它。
- 在 macOS 上,可以在 "应用程序" 文件夹中找到 Anaconda-Navigator,点击启动。
在 Anaconda 环境中进行配置之前,首先需要将你的 GitHub 仓库克隆到本地。
- 打开终端(macOS)或 Git Bash(Windows)。
- 运行以下命令,克隆你的仓库:
git clone [email protected]:itWangCode/AI-drug-design.git这将下载包含 AIDrugDesign.yml 文件的仓库到本地。
进入克隆的项目文件夹,并通过 .yml 文件创建新的 Conda 环境:
- 在终端或 Git Bash 中,切换到你克隆的仓库目录:
cd AI-drug-design- 运行以下命令,根据
AIDrugDesign.yml文件创建 Conda 环境:
conda env create -f AIDrugDesign.yml❤️请您耐心等待15分钟以上,请您连接 wifi !!!!!!!
- Conda 将自动根据
.yml文件安装所需的依赖包并创建环境。
安装完成后,使用以下命令查看所有环境,检查是否创建了 AI-drug-design 环境:
conda env list你应该会看到类似如下的输出,其中包含 AI-drug-design:
# conda environments:
#
base * /path/to/anaconda3
AI-drug-design /path/to/anaconda3/envs/AI-drug-design最后,使用以下命令激活 AI-drug-design 环境:
conda activate AI-drug-design你现在已成功配置并激活了 AI-drug-design 环境,可以开始使用该环境进行开发了。
我有时候特别怕Macos系统同学
Mac系统 PyCharm.app”已损坏,无法打开。 您应该将它移到废纸篓。 mac系统下载好了pycharm后提示,Mac系统 PyCharm.app”已损坏,无法打开。 您应该将它移到废纸篓。 我们在terminal窗口输入一下命令后就可以正常打开软件了。前提你的pycharm在你的应用程序文件夹内。 sudo xattr -r -d com.apple.quarantine /Applications/PyCharm.app
废话不多说,先上 Pycharm 2024.2.1 版本破解成功的截图,如下,可以看到已经成功破解到 2099 年辣,舒服!
接下来,我就将通过图文的方式, 来详细讲解如何激活 Pycharm 2024.2.1 版本至 2099 年。首先,如果小伙伴的电脑上有安装老版本的 Pycharm , 需要将其卸载掉,如下所示(没有安装则不用管,直接安装即可):
将删除缓存和本地历史勾选上,点击卸载按钮开始卸载:
卸载完成后,点击关闭按钮。
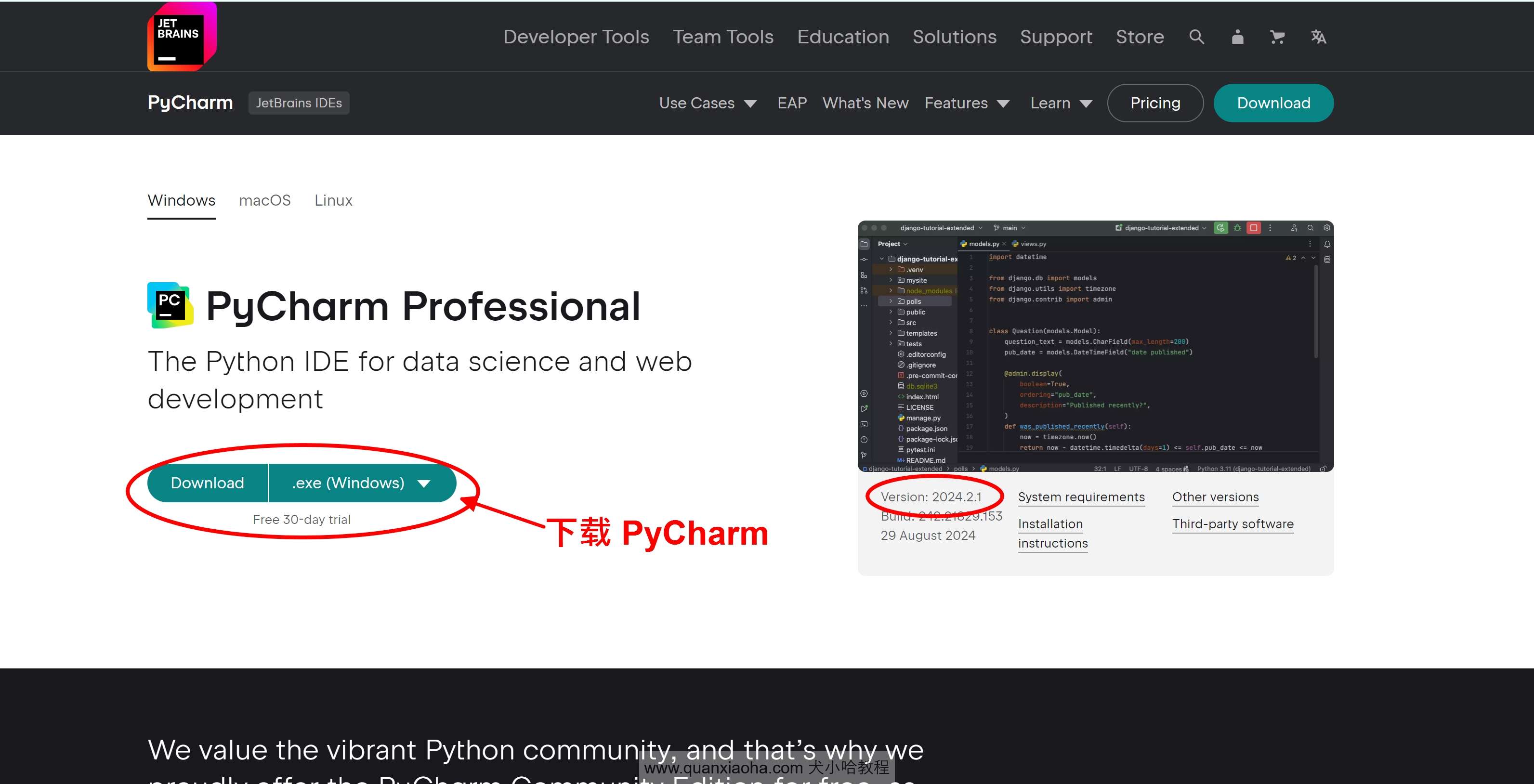
访问 Pycharm 官网:https://www.jetbrains.com/pycharm/download,下载 Pycharm 2024.2.1 版本的安装包。
下载完成后,双击 .exe 安装包开始安装 Pycharm :
点击下一步按钮:
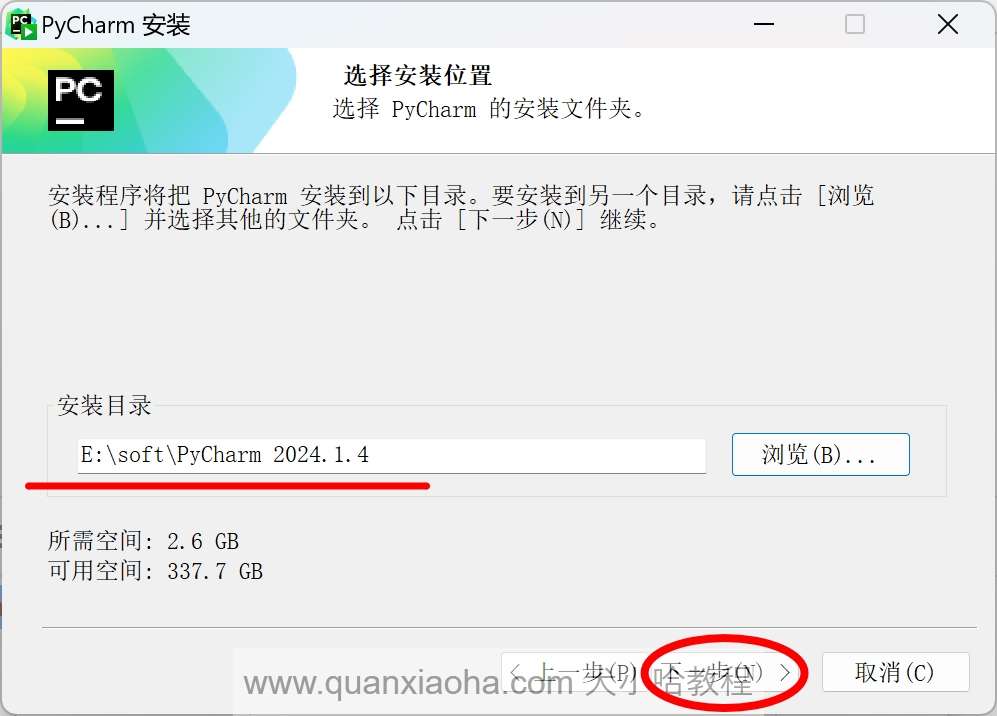
自定义安装路径,我这里安装在了 E:\ 盘下,继续点击下一步按钮:
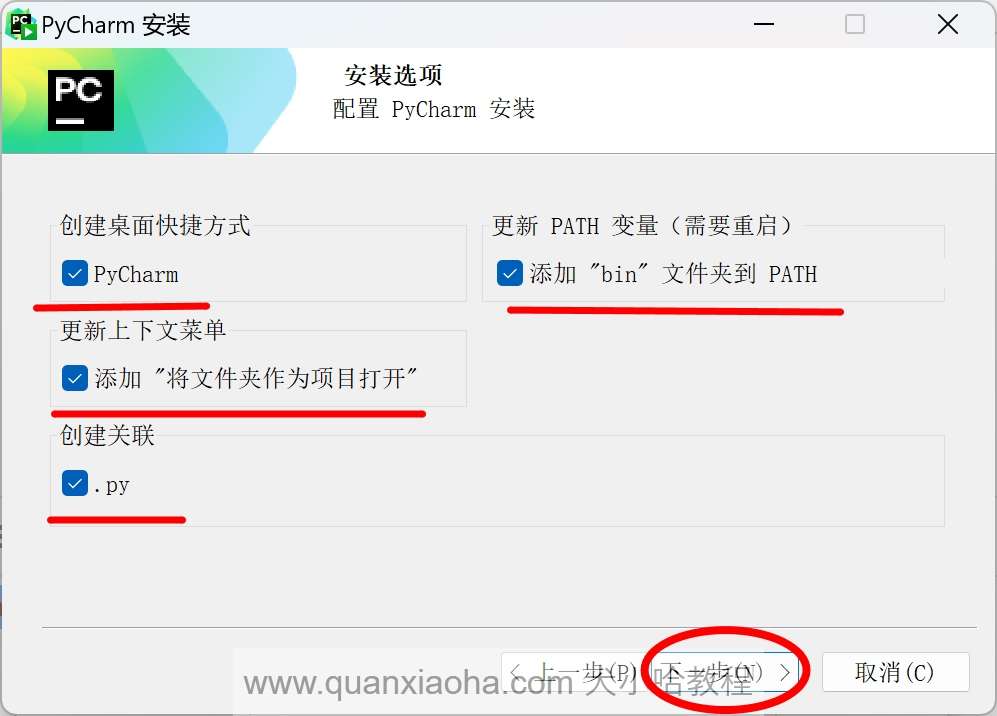
进入到安装选项的选择,将下图标注的部分,全部勾选上,点击下一步按钮:
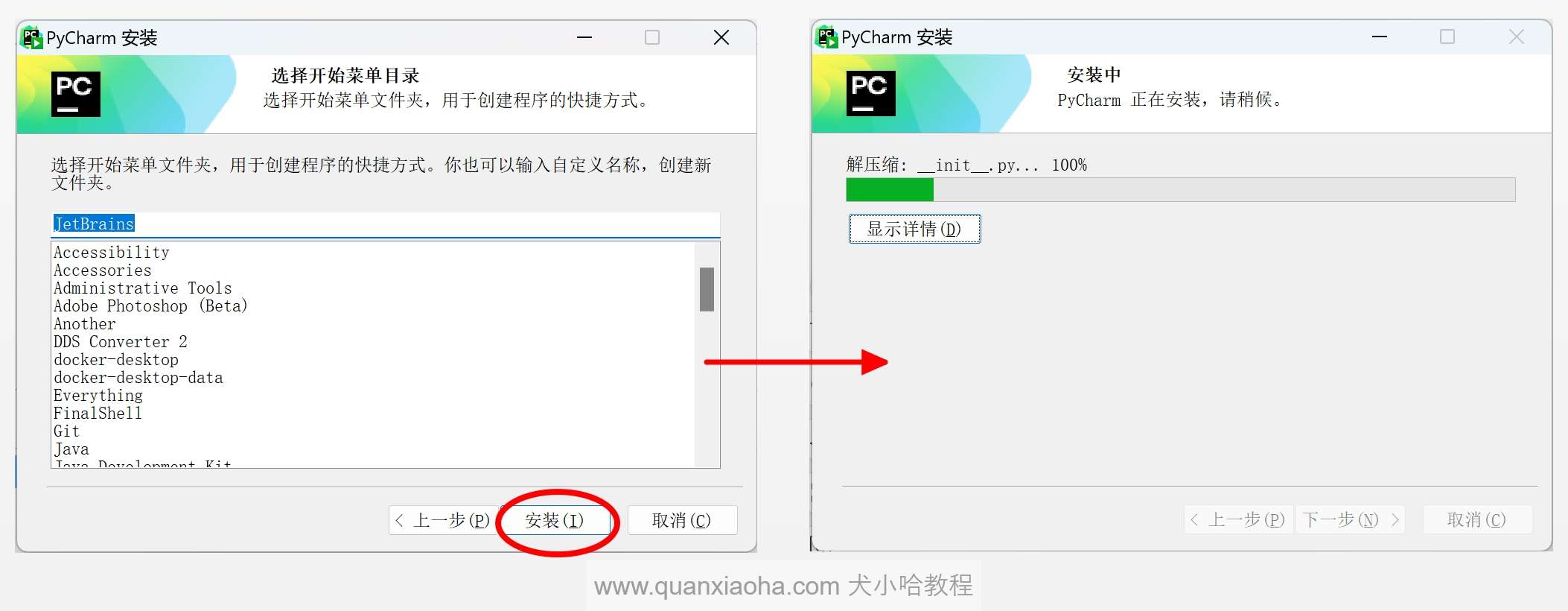
点击安装按钮,等待 Pycharm 安装完成:
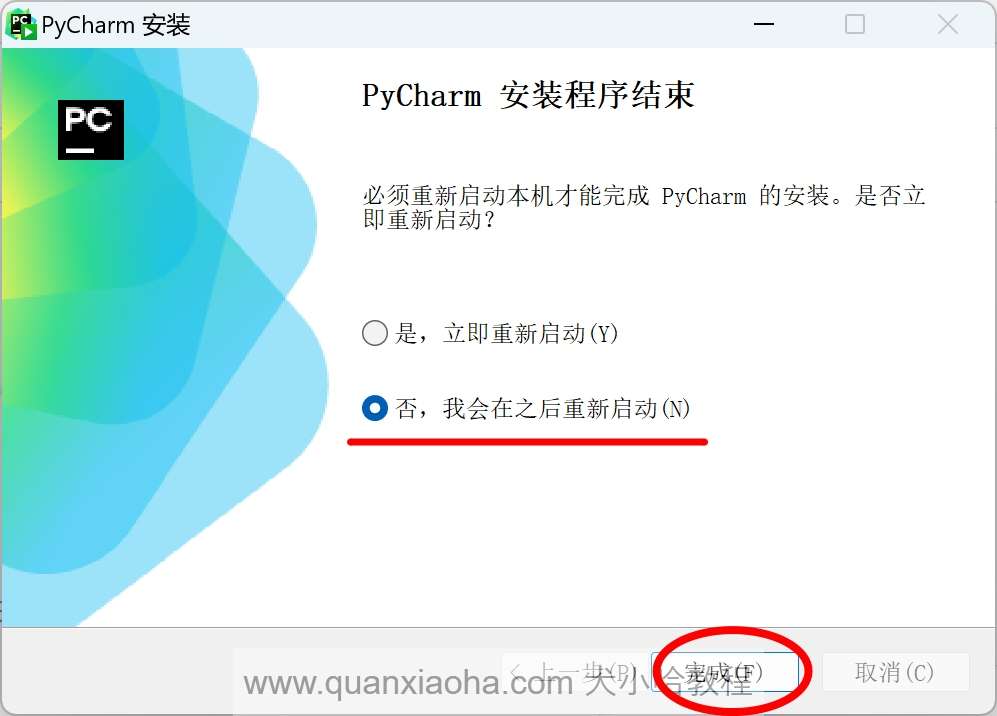
安装成功后,会弹出如下提示框,一个是立即启动,不要勾选它。我们勾选 “否,我会在之后重新启动”,因为需要先破解成功后再启动 Pycharm :
点击完成按钮,关闭弹框。
破解脚本我放置在了网盘中,并提供了多个备用链接,以防下载失效。
**提示:破解脚本的网盘链接文末获取 ~**
**提示:破解脚本的网盘链接文末获取 ~**
**提示:破解脚本的网盘链接文末获取 ~**
下载成功后,如下,是个压缩包,先对它进行解压:
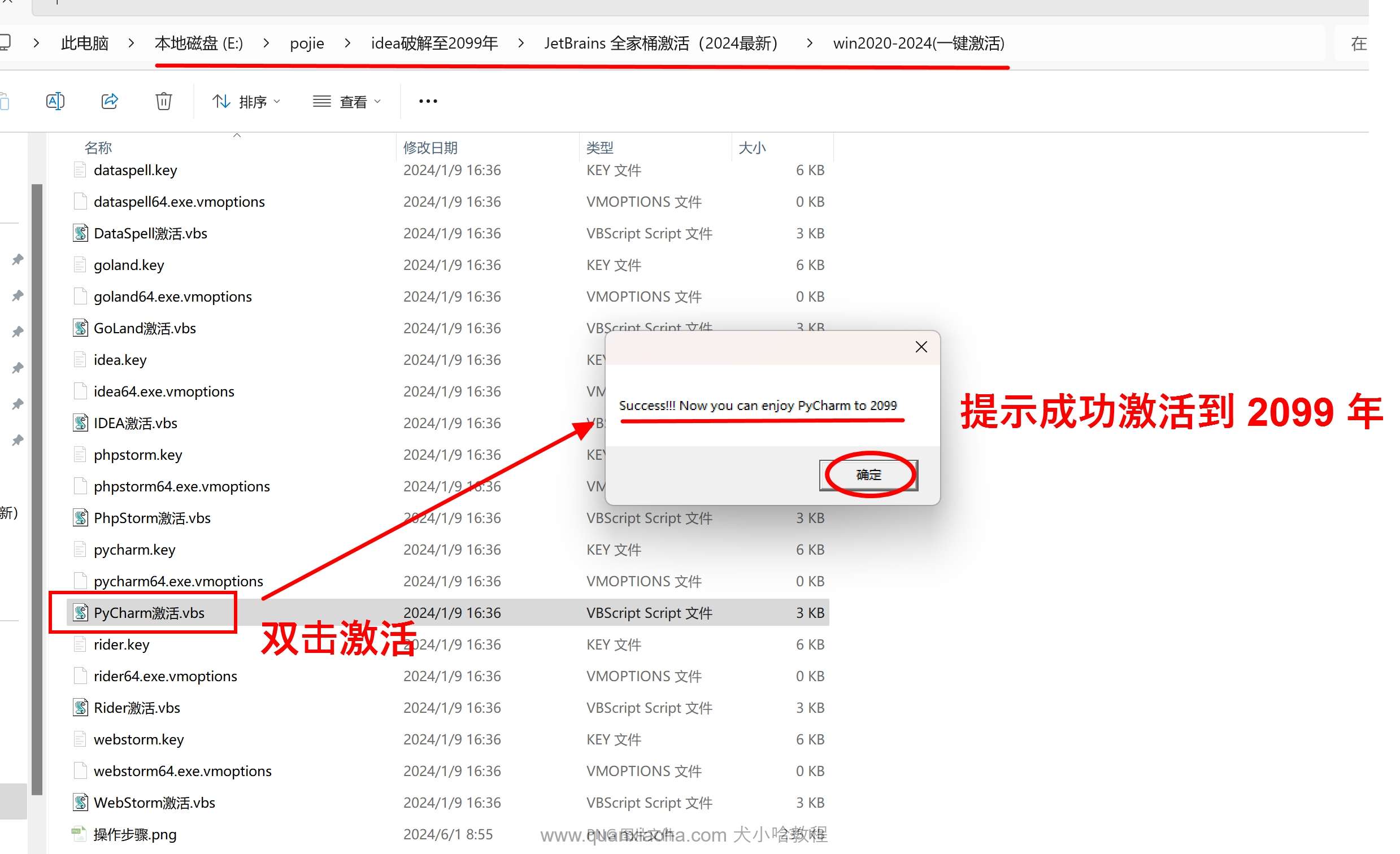
进入到解压后的文件夹 /win2020-2024(一键激活) 中,双击 Pycharm 激活.vbs ,若提示 Success!!! Now you can enjoy Pycharm to 2099 , 则表示 Pycharm 激活成功啦 ~
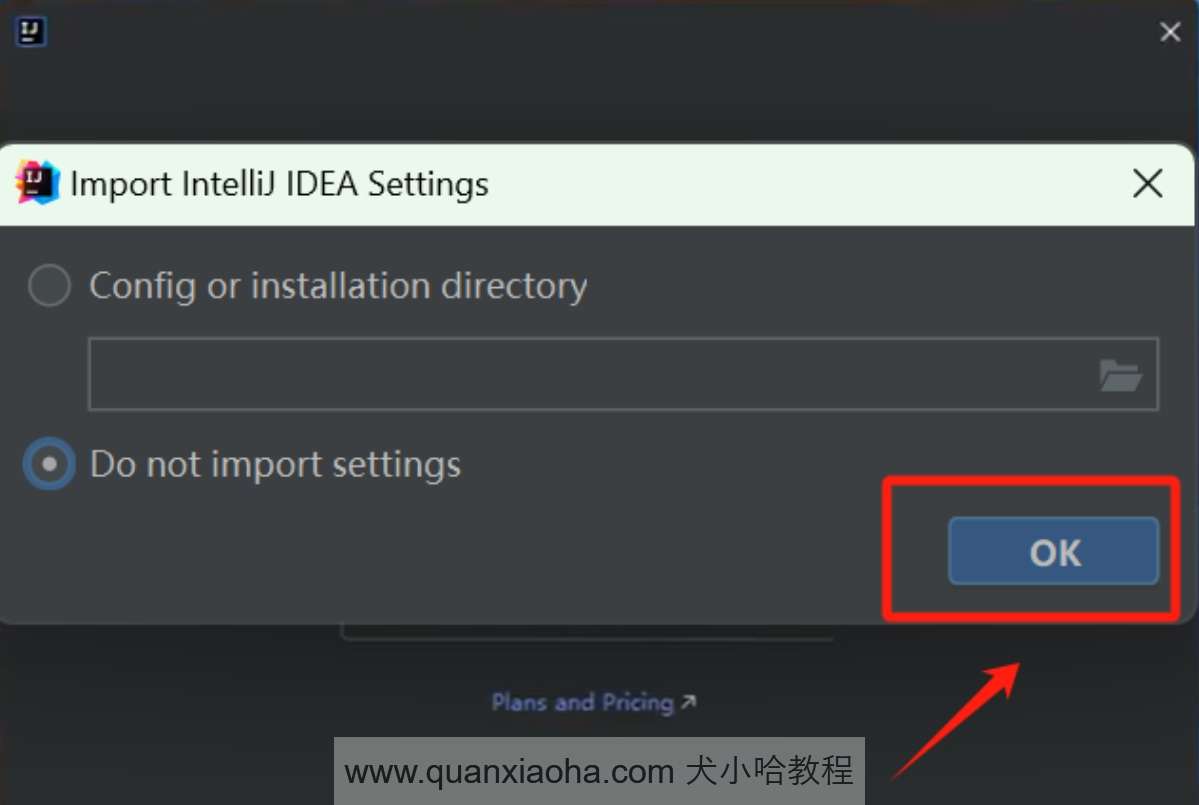
激活成功后,双击桌面的 Pycharm 快捷启动图标,来打开 Pycharm 。注意,首次安装 Pycharm 可能会弹出如下提示框,勾选 Do not import settings, 点击 OK 按钮即可:
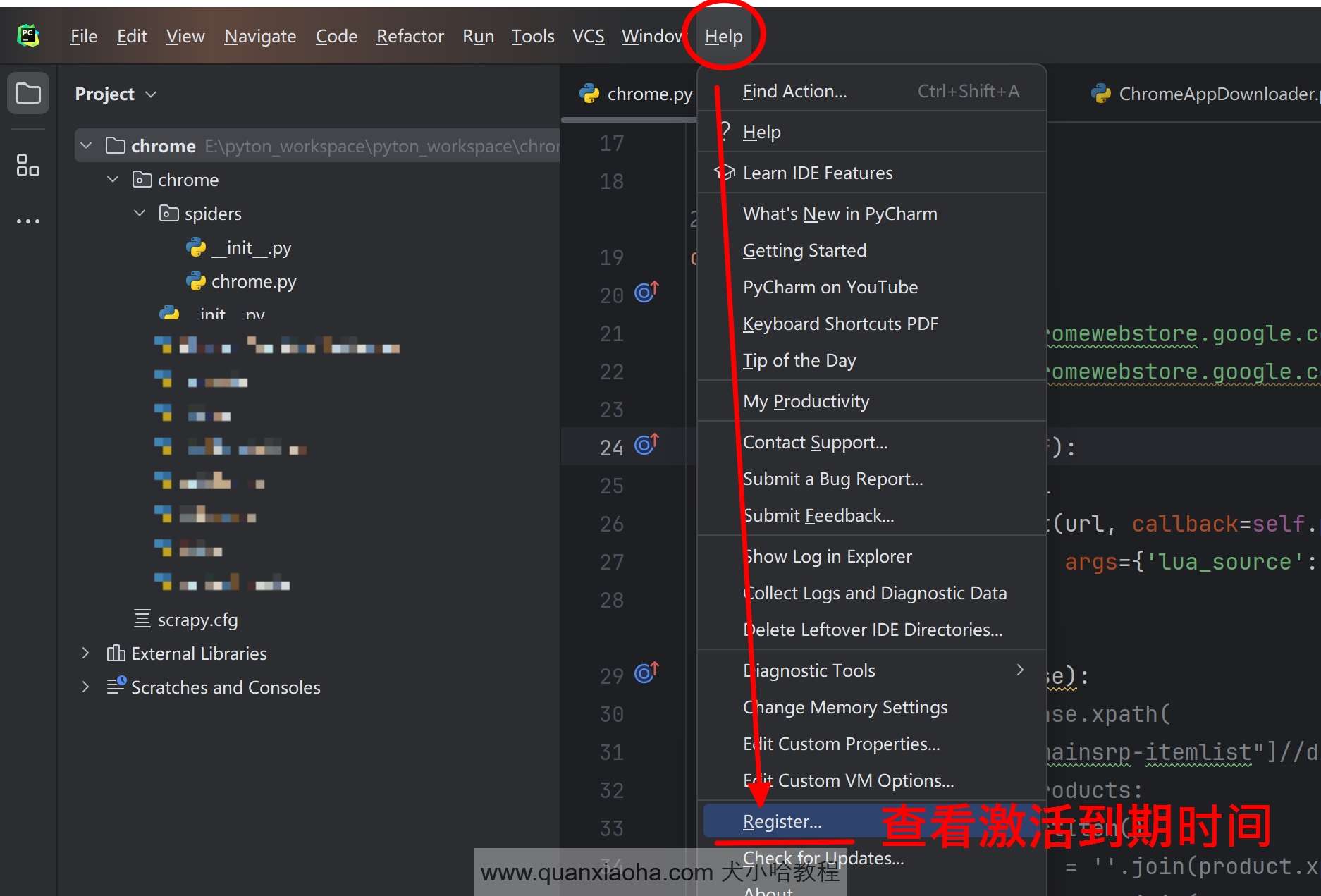
进入 Pycharm 中后,点击菜单栏 Help | Register... , 即可查看 Pycharm 的激活到期时间:
EUWT4EE9X2-eyJsaWNlbnNlSWQiOiJFVVdUNEVFOVgyIiwibGljZW5zZWVOYW1lIjoic2lnbnVwIHNjb290ZXIiLCJhc3NpZ25lZU5hbWUiOiIiLCJhc3NpZ25lZUVtYWlsIjoiIiwibGljZW5zZVJlc3RyaWN0aW9uIjoiIiwiY2hlY2tDb25jdXJyZW50VXNlIjpmYWxzZSwicHJvZHVjdHMiOlt7ImNvZGUiOiJQU0kiLCJmYWxsYmFja0RhdGUiOiIyMDI1LTA4LTAxIiwicGFpZFVwVG8iOiIyMDI1LTA4LTAxIiwiZXh0ZW5kZWQiOnRydWV9LHsiY29kZSI6IlBDIiwiZmFsbGJhY2tEYXRlIjoiMjAyNS0wOC0wMSIsInBhaWRVcFRvIjoiMjAyNS0wOC0wMSIsImV4dGVuZGVkIjpmYWxzZX0seyJjb2RlIjoiUFBDIiwiZmFsbGJhY2tEYXRlIjoiMjAyNS0wOC0wMSIsInBhaWRVcFRvIjoiMjAyNS0wOC0wMSIsImV4dGVuZGVkIjp0cnVlfSx7ImNvZGUiOiJQV1MiLCJmYWxsYmFja0RhdGUiOiIyMDI1LTA4LTAxIiwicGFpZFVwVG8iOiIyMDI1LTA4LTAxIiwiZXh0ZW5kZWQiOnRydWV9LHsiY29kZSI6IlBDV01QIiwiZmFsbGJhY2tEYXRlIjoiMjAyNS0wOC0wMSIsInBhaWRVcFRvIjoiMjAyNS0wOC0wMSIsImV4dGVuZGVkIjp0cnVlfV0sIm1ldGFkYXRhIjoiMDEyMDIyMDkwMlBTQU4wMDAwMDUiLCJoYXNoIjoiVFJJQUw6MzUzOTQ0NTE3IiwiZ3JhY2VQZXJpb2REYXlzIjo3LCJhdXRvUHJvbG9uZ2F0ZWQiOmZhbHNlLCJpc0F1dG9Qcm9sb25nYXRlZCI6ZmFsc2V9-FT9l1nyyF9EyNmlelrLP9rGtugZ6sEs3CkYIKqGgSi608LIamge623nLLjI8f6O4EdbCfjJcPXLxklUe1O/5ASO3JnbPFUBYUEebCWZPgPfIdjw7hfA1PsGUdw1SBvh4BEWCMVVJWVtc9ktE+gQ8ldugYjXs0s34xaWjjfolJn2V4f4lnnCv0pikF7Ig/Bsyd/8bsySBJ54Uy9dkEsBUFJzqYSfR7Z/xsrACGFgq96ZsifnAnnOvfGbRX8Q8IIu0zDbNh7smxOwrz2odmL72UaU51A5YaOcPSXRM9uyqCnSp/ENLzkQa/B9RNO+VA7kCsj3MlJWJp5Sotn5spyV+gA==-MIIETDCCAjSgAwIBAgIBDTANBgkqhkiG9w0BAQsFADAYMRYwFAYDVQQDDA1KZXRQcm9maWxlIENBMB4XDTIwMTAxOTA5MDU1M1oXDTIyMTAyMTA5MDU1M1owHzEdMBsGA1UEAwwUcHJvZDJ5LWZyb20tMjAyMDEwMTkwggEiMA0GCSqGSIb3DQEBAQUAA4IBDwAwggEKAoIBAQCUlaUFc1wf+CfY9wzFWEL2euKQ5nswqb57V8QZG7d7RoR6rwYUIXseTOAFq210oMEe++LCjzKDuqwDfsyhgDNTgZBPAaC4vUU2oy+XR+Fq8nBixWIsH668HeOnRK6RRhsr0rJzRB95aZ3EAPzBuQ2qPaNGm17pAX0Rd6MPRgjp75IWwI9eA6aMEdPQEVN7uyOtM5zSsjoj79Lbu1fjShOnQZuJcsV8tqnayeFkNzv2LTOlofU/Tbx502Ro073gGjoeRzNvrynAP03pL486P3KCAyiNPhDs2z8/COMrxRlZW5mfzo0xsK0dQGNH3UoG/9RVwHG4eS8LFpMTR9oetHZBAgMBAAGjgZkwgZYwCQYDVR0TBAIwADAdBgNVHQ4EFgQUJNoRIpb1hUHAk0foMSNM9MCEAv8wSAYDVR0jBEEwP4AUo562SGdCEjZBvW3gubSgUouX8bOhHKQaMBgxFjAUBgNVBAMMDUpldFByb2ZpbGUgQ0GCCQDSbLGDsoN54TATBgNVHSUEDDAKBggrBgEFBQcDATALBgNVHQ8EBAMCBaAwDQYJKoZIhvcNAQELBQADggIBABqRoNGxAQct9dQUFK8xqhiZaYPd30TlmCmSAaGJ0eBpvkVeqA2jGYhAQRqFiAlFC63JKvWvRZO1iRuWCEfUMkdqQ9VQPXziE/BlsOIgrL6RlJfuFcEZ8TK3syIfIGQZNCxYhLLUuet2HE6LJYPQ5c0jH4kDooRpcVZ4rBxNwddpctUO2te9UU5/FjhioZQsPvd92qOTsV+8Cyl2fvNhNKD1Uu9ff5AkVIQn4JU23ozdB/R5oUlebwaTE6WZNBs+TA/qPj+5/we9NH71WRB0hqUoLI2AKKyiPw++FtN4Su1vsdDlrAzDj9ILjpjJKA1ImuVcG329/WTYIKysZ1CWK3zATg9BeCUPAV1pQy8ToXOq+RSYen6winZ2OO93eyHv2Iw5kbn1dqfBw1BuTE29V2FJKicJSu8iEOpfoafwJISXmz1wnnWL3V/0NxTulfWsXugOoLfv0ZIBP1xH9kmf22jjQ2JiHhQZP7ZDsreRrOeIQ/c4yR8IQvMLfC0WKQqrHu5ZzXTH4NO3CwGWSlTY74kE91zXB5mwWAx1jig+UXYc2w4RkVhy0//lOmVya/PEepuuTTI4+UJwC7qbVlh5zfhj8oTNUXgN0AOc+Q0/WFPl1aw5VV/VrO8FCoB15lFVlpKaQ1Yh+DVU8ke+rt9Th0BCHXe0uZOEmH0nOnH/0onD
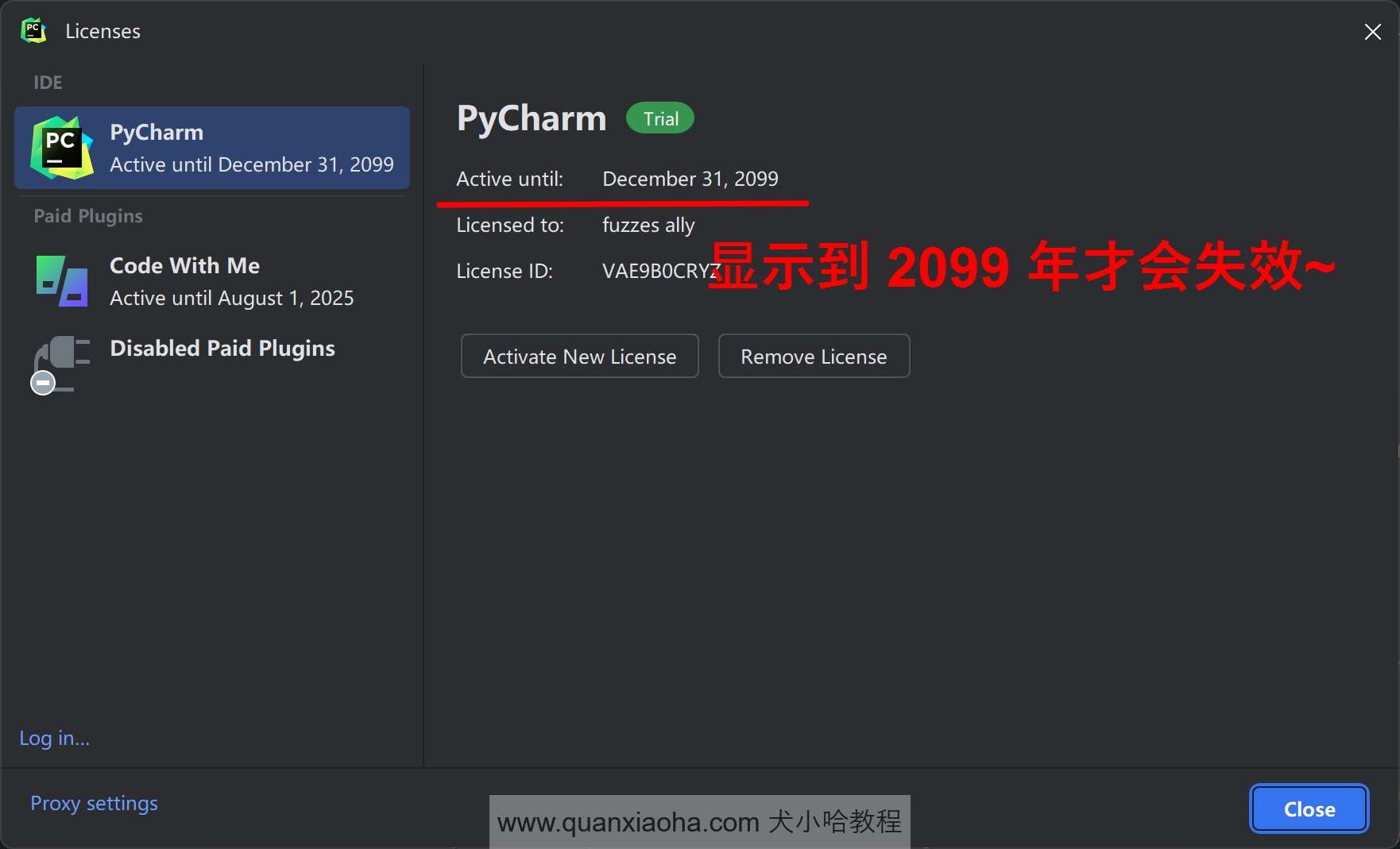
如下图标注所示,根据日期显示直到 2099 年才会失效,确实是破解成功了:
PS: 激活脚本由于提取人数过多,导致分享的百度网盘链接容易被封:
蛋疼 ing,为限制人数,目前暂不提供页面直接下载,改为从笔者公众号下载。
需要的小伙伴,扫描下方公众号二维码,或者关注公众号: Python 见习室,回复关键字:2099, 即可免费无套路获取激活码、破解补丁,持续更新中 ~。
更多的额操作:请您参考:【https://www.exception.site/article/1762】
在学习 Python 用于 AI 药物设计(AI-drug-design)项目时,您需要掌握以下知识点。学习计划将涵盖基础知识、数据处理、机器学习、深度学习框架、药物设计相关库的使用等内容。以下是详细的学习目录和教案:
-
Python 基础
1.1 Python 环境配置(Anaconda、虚拟环境、Jupyter Notebook) 1.2 Python 基础语法(变量、数据类型、运算符) 1.3 控制结构(条件语句、循环) 1.4 函数与模块(自定义函数、导入库) 1.5 文件操作(读取和写入文件) 1.6 异常处理 -
数据处理与分析
2.1 Numpy 数组操作 2.2 Pandas 数据框操作 2.3 数据清洗与处理 2.4 数据可视化(Matplotlib、Seaborn) 2.5 基础统计与数据分析方法 -
科学计算与机器学习基础
3.1 Scikit-learn 入门 3.2 常用算法(线性回归、分类、聚类、决策树等) 3.3 模型评估与优化 3.4 模型调参与交叉验证 -
深度学习框架
4.1 TensorFlow 基础 4.2 Keras 快速上手 4.3 PyTorch 基础 4.4 GPU 加速与优化 4.5 神经网络的构建与训练 -
AI 药物设计基础
5.1 药物设计相关的 Python 库介绍 5.2 RDKit(化学信息学工具)入门与使用 5.3 Mol2Vec 分子特征表示方法 5.4 化合物的预处理与分子特征提取 5.5 分子对接与虚拟筛选 5.6 药物活性预测模型 -
高级主题
6.1 深度学习在药物设计中的应用 6.2 分子生成模型(GAN、VAE) 6.3 分子动力学模拟简介 6.4 蛋白质结构预测与分子对接 6.5 AI 在药物筛选和优化中的应用
目标:掌握 Python 基本语法、数据结构及控制流,能够编写简单的 Python 程序。
教学内容:
- Python 环境配置与解释器运行
- 基本数据类型(字符串、整数、浮点数、布尔)
- 数据结构:列表、字典、集合、元组
- 控制结构:if、else、for、while 循环
- 函数与参数传递,理解递归
- 模块的导入与创建(如
math、random模块)
作业:
- 编写一个处理简单文本的 Python 程序(如计算文本单词频率)
- 编写一个函数,接受多个参数并返回最大值
目标:学习 Numpy 和 Pandas 库,能够进行高效的数据处理和分析。
教学内容:
- Numpy 基础:数组创建、形状修改、索引与切片、数组运算
- Pandas 基础:Series 和 DataFrame 的操作,缺失值处理,数据筛选与排序
- 数据的导入与导出(CSV、Excel 等格式)
- 数据可视化:柱状图、折线图、散点图
- 数据统计分析:均值、中位数、标准差等
作业:
- 使用 Pandas 读取 CSV 文件,计算每列的均值和标准差
- 使用 Matplotlib 绘制简单的柱状图和折线图
目标:掌握机器学习基础理论,能够使用 Scikit-learn 进行模型训练和评估。
教学内容:
- 监督学习和无监督学习简介
- 数据集的划分:训练集、验证集与测试集
- 线性回归、逻辑回归、KNN、决策树等基本算法
- 模型的评估:准确率、混淆矩阵、ROC 曲线
- 超参数优化与交叉验证
作业:
- 使用 Scikit-learn 进行一个简单的分类问题(如鸢尾花数据集分类)
- 绘制模型的 ROC 曲线并计算 AUC
目标:熟悉常用的深度学习框架(TensorFlow、Keras、PyTorch),能够搭建简单的神经网络。
教学内容:
- TensorFlow 和 Keras 的基本用法:张量操作、模型构建
- PyTorch 的基本操作:Tensors、Autograd、优化器
- 构建简单的全连接神经网络
- 使用 GPU 进行模型训练
- 优化方法:SGD、Adam 等优化器
- 避免过拟合的正则化方法(如 Dropout)
作业:
- 使用 Keras 实现一个手写数字识别模型(MNIST 数据集)
- 使用 PyTorch 实现一个简单的卷积神经网络
目标:学习 AI 药物设计相关的库,能够进行分子数据的处理与建模。
教学内容:
- RDKit 入门:分子结构的读取、绘制与操作
- 化合物分子描述符的计算
- 化学库的虚拟筛选
- Mol2Vec 特征表示方法
- 基于机器学习的药物活性预测模型
- 小分子的对接模拟(AutoDock、PyMOL 简介)
作业:
- 使用 RDKit 对一组化合物进行特征提取
- 使用机器学习模型预测药物活性
目标:掌握深度学习在分子生成与药物设计中的应用。
教学内容:
- 分子生成模型:生成对抗网络(GAN)、变分自编码器(VAE)
- 基于深度学习的分子优化方法
- 蛋白质结构预测:AlphaFold 介绍
- 分子动力学模拟基础与应用
- 药物筛选流程与 AI 在其中的应用
作业:
- 使用 GAN 模型生成新的分子结构
- 编写脚本对某一蛋白质靶点进行分子对接模拟
For Tasks:
Click tags to check more tools for each tasksFor Jobs:
Alternative AI tools for AI-Drug-Discovery-Design
Similar Open Source Tools

AI-Drug-Discovery-Design
AI-Drug-Discovery-Design is a repository focused on Artificial Intelligence-assisted Drug Discovery and Design. It explores the use of AI technology to accelerate and optimize the drug development process. The advantages of AI in drug design include speeding up research cycles, improving accuracy through data-driven models, reducing costs by minimizing experimental redundancies, and enabling personalized drug design for specific patients or disease characteristics.
LLMAI-writer
LLMAI-writer is a powerful AI tool for assisting in novel writing, utilizing state-of-the-art large language models to help writers brainstorm, plan, and create novels. Whether you are an experienced writer or a beginner, LLMAI-writer can help you efficiently complete the writing process.
MaiMBot
MaiMBot is an intelligent QQ group chat bot based on a large language model. It is developed using the nonebot2 framework, utilizes LLM for conversation abilities, MongoDB for data persistence, and NapCat for QQ protocol support. The bot features keyword-triggered proactive responses, dynamic prompt construction, support for images and message forwarding, typo generation, multiple replies, emotion-based emoji responses, daily schedule generation, user relationship management, knowledge base, and group impressions. Work-in-progress features include personality, group atmosphere, image handling, humor, meme functions, and Minecraft interactions. The tool is in active development with plans for GIF compatibility, mini-program link parsing, bug fixes, documentation improvements, and logic enhancements for emoji sending.
AHU-AI-Repository
This repository is dedicated to the learning and exchange of resources for the School of Artificial Intelligence at Anhui University. Notes will be published on this website first: https://www.aoaoaoao.cn and will be synchronized to the repository regularly. You can also contact me at [email protected].
claude-init
Claude Code Chinese development suite is a localized version based on the Claude Code Development Kit, offering a seamless Chinese AI programming experience. It features complete Chinese AI commands, documentation system, error messages, and installation experience. The suite includes intelligent context management with a three-tier document structure, automatic context injection, smart document routing, and cross-session state management. It integrates development tools like Hook system, MCP server support, security scans, and notification system. Additionally, it provides a comprehensive template library with project templates, document templates, and configuration examples.
godoos
GodoOS is an efficient intranet office operating system that includes various office tools such as word/excel/ppt/pdf/internal chat/whiteboard/mind map, with native file storage support. The platform interface mimics the Windows style, making it easy to operate while maintaining low resource consumption and high performance. It automatically connects to intranet users without registration, enabling instant communication and file sharing. The flexible and highly configurable app store allows for unlimited expansion.
MoneyPrinterTurbo
MoneyPrinterTurbo is a tool that can automatically generate video content based on a provided theme or keyword. It can create video scripts, materials, subtitles, and background music, and then compile them into a high-definition short video. The tool features a web interface and an API interface, supporting AI-generated video scripts, customizable scripts, multiple HD video sizes, batch video generation, customizable video segment duration, multilingual video scripts, multiple voice synthesis options, subtitle generation with font customization, background music selection, access to high-definition and copyright-free video materials, and integration with various AI models like OpenAI, moonshot, Azure, and more. The tool aims to simplify the video creation process and offers future plans to enhance voice synthesis, add video transition effects, provide more video material sources, offer video length options, include free network proxies, enable real-time voice and music previews, support additional voice synthesis services, and facilitate automatic uploads to YouTube platform.
DocTranslator
DocTranslator is a document translation tool that supports various file formats, compatible with OpenAI format API, and offers batch operations and multi-threading support. Whether for individual users or enterprise teams, DocTranslator helps efficiently complete document translation tasks. It supports formats like txt, markdown, word, csv, excel, pdf (non-scanned), and ppt for AI translation. The tool is deployed using Docker for easy setup and usage.
Con-Nav-Item
Con-Nav-Item is a modern personal navigation system designed for digital workers. It is not just a link bookmark but also an all-in-one workspace integrated with AI smart generation, multi-device synchronization, card-based management, and deep browser integration.
CodeAsk
CodeAsk is a code analysis tool designed to tackle complex issues such as code that seems to self-replicate, cryptic comments left by predecessors, messy and unclear code, and long-lasting temporary solutions. It offers intelligent code organization and analysis, security vulnerability detection, code quality assessment, and other interesting prompts to help users understand and work with legacy code more efficiently. The tool aims to translate 'legacy code mountains' into understandable language, creating an illusion of comprehension and facilitating knowledge transfer to new team members.
bk-lite
Blueking Lite is an AI First lightweight operation product with low deployment resource requirements, low usage costs, and progressive experience, providing essential tools for operation administrators.
MarkMap-OpenAi-ChatGpt
MarkMap-OpenAi-ChatGpt is a Vue.js-based mind map generation tool that allows users to generate mind maps by entering titles or content. The application integrates the markmap-lib and markmap-view libraries, supports visualizing mind maps, and provides functions for zooming and adapting the map to the screen. Users can also export the generated mind map in PNG, SVG, JPEG, and other formats. This project is suitable for quickly organizing ideas, study notes, project planning, etc. By simply entering content, users can get an intuitive mind map that can be continuously expanded, downloaded, and shared.

LabelQuick
LabelQuick_V2.0 is a fast image annotation tool designed and developed by the AI Horizon team. This version has been optimized and improved based on the previous version. It provides an intuitive interface and powerful annotation and segmentation functions to efficiently complete dataset annotation work. The tool supports video object tracking annotation, quick annotation by clicking, and various video operations. It introduces the SAM2 model for accurate and efficient object detection in video frames, reducing manual intervention and improving annotation quality. The tool is designed for Windows systems and requires a minimum of 6GB of memory.
Snap-Solver
Snap-Solver is a revolutionary AI tool for online exam solving, designed for students, test-takers, and self-learners. With just a keystroke, it automatically captures any question on the screen, analyzes it using AI, and provides detailed answers. Whether it's complex math formulas, physics problems, coding issues, or challenges from other disciplines, Snap-Solver offers clear, accurate, and structured solutions to help you better understand and master the subject matter.
prompt-optimizer
Prompt Optimizer is a powerful AI prompt optimization tool that helps you write better AI prompts, improving AI output quality. It supports both web application and Chrome extension usage. The tool features intelligent optimization for prompt words, real-time testing to compare before and after optimization, integration with multiple mainstream AI models, client-side processing for security, encrypted local storage for data privacy, responsive design for user experience, and more.
llm-resource
llm-resource is a comprehensive collection of high-quality resources for Large Language Models (LLM). It covers various aspects of LLM including algorithms, training, fine-tuning, alignment, inference, data engineering, compression, evaluation, prompt engineering, AI frameworks, AI basics, AI infrastructure, AI compilers, LLM application development, LLM operations, AI systems, and practical implementations. The repository aims to gather and share valuable resources related to LLM for the community to benefit from.
For similar tasks
AI2BMD
AI2BMD is a program for efficiently simulating protein molecular dynamics with ab initio accuracy. The repository contains datasets, simulation programs, and public materials related to AI2BMD. It provides a Docker image for easy deployment and a standalone launcher program. Users can run simulations by downloading the launcher script and specifying simulation parameters. The repository also includes ready-to-use protein structures for testing. AI2BMD is designed for x86-64 GNU/Linux systems with recommended hardware specifications. The related research includes model architectures like ViSNet, Geoformer, and fine-grained force metrics for MLFF. Citation information and contact details for the AI2BMD Team are provided.

AI-Drug-Discovery-Design
AI-Drug-Discovery-Design is a repository focused on Artificial Intelligence-assisted Drug Discovery and Design. It explores the use of AI technology to accelerate and optimize the drug development process. The advantages of AI in drug design include speeding up research cycles, improving accuracy through data-driven models, reducing costs by minimizing experimental redundancies, and enabling personalized drug design for specific patients or disease characteristics.
For similar jobs

AlphaFold3
AlphaFold3 is an implementation of the Alpha Fold 3 model in PyTorch for accurate structure prediction of biomolecular interactions. It includes modules for genetic diffusion and full model examples for forward pass computations. The tool allows users to generate random pair and single representations, operate on atomic coordinates, and perform structure predictions based on input tensors. The implementation also provides functionalities for training and evaluating the model.
biochatter
Generative AI models have shown tremendous usefulness in increasing accessibility and automation of a wide range of tasks. This repository contains the `biochatter` Python package, a generic backend library for the connection of biomedical applications to conversational AI. It aims to provide a common framework for deploying, testing, and evaluating diverse models and auxiliary technologies in the biomedical domain. BioChatter is part of the BioCypher ecosystem, connecting natively to BioCypher knowledge graphs.
admet_ai
ADMET-AI is a platform for ADMET prediction using Chemprop-RDKit models trained on ADMET datasets from the Therapeutics Data Commons. It offers command line, Python API, and web server interfaces for making ADMET predictions on new molecules. The platform can be easily installed using pip and supports GPU acceleration. It also provides options for processing TDC data, plotting results, and hosting a web server. ADMET-AI is a machine learning platform for evaluating large-scale chemical libraries.

AI-Drug-Discovery-Design
AI-Drug-Discovery-Design is a repository focused on Artificial Intelligence-assisted Drug Discovery and Design. It explores the use of AI technology to accelerate and optimize the drug development process. The advantages of AI in drug design include speeding up research cycles, improving accuracy through data-driven models, reducing costs by minimizing experimental redundancies, and enabling personalized drug design for specific patients or disease characteristics.
bionemo-framework
NVIDIA BioNeMo Framework is a collection of programming tools, libraries, and models for computational drug discovery. It accelerates building and adapting biomolecular AI models by providing domain-specific, optimized models and tooling for GPU-based computational resources. The framework offers comprehensive documentation and support for both community and enterprise users.
New-AI-Drug-Discovery
New AI Drug Discovery is a repository focused on the applications of Large Language Models (LLM) in drug discovery. It provides resources, tools, and examples for leveraging LLM technology in the pharmaceutical industry. The repository aims to showcase the potential of using AI-driven approaches to accelerate the drug discovery process, improve target identification, and optimize molecular design. By exploring the intersection of artificial intelligence and drug development, this repository offers insights into the latest advancements in computational biology and cheminformatics.
gromacs_copilot
GROMACS Copilot is an agent designed to automate molecular dynamics simulations for proteins in water using GROMACS. It handles system setup, simulation execution, and result analysis automatically, providing outputs such as RMSD, RMSF, Rg, and H-bonds. Users can interact with the agent through prompts and API keys from DeepSeek and OpenAI. The tool aims to simplify the process of running MD simulations, allowing users to focus on other tasks while it handles the technical aspects of the simulations.

Biomni
Biomni is a general-purpose biomedical AI agent designed to autonomously execute a wide range of research tasks across diverse biomedical subfields. By integrating cutting-edge large language model (LLM) reasoning with retrieval-augmented planning and code-based execution, Biomni helps scientists dramatically enhance research productivity and generate testable hypotheses.