Best AI tools for< Cell Biologist >
Infographic
4 - AI tool Sites
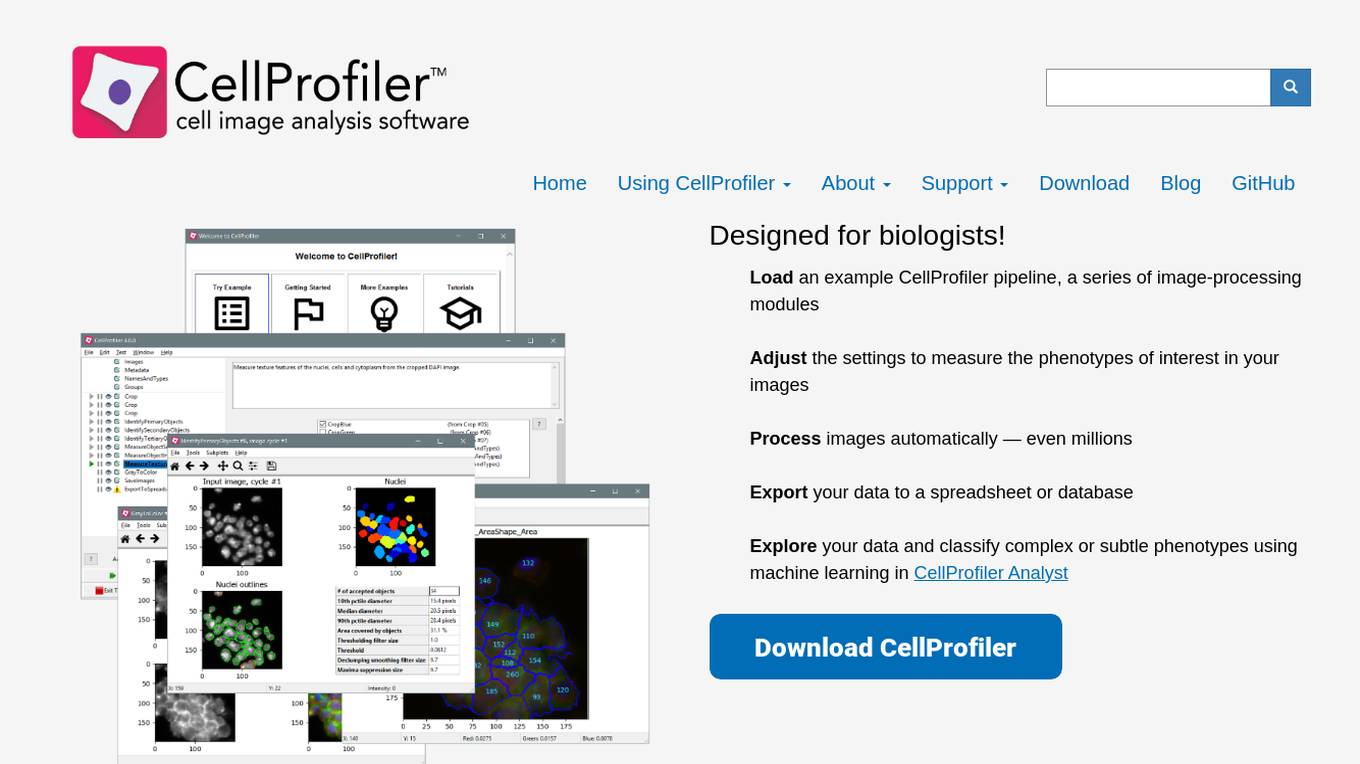
CellProfiler
CellProfiler is an AI tool designed for biologists to analyze and process images automatically. It allows users to load image-processing modules, adjust settings, measure phenotypes, export data, and classify phenotypes using machine learning. The application is user-friendly and provides a seamless experience for biologists to analyze complex or subtle phenotypes in their images.
Deepcell
Deepcell is a company that develops technology for single-cell analysis. Their REM-I platform combines label-free imaging, deep learning, and gentle sorting to leverage single cell morphology as a high-dimensional quantitative readout. This allows researchers to gain insights into cells' phenotype and function to address important research questions across biology.
Byterat
Byterat is a cloud-based platform that provides battery data management, visualization, and analytics. It offers an end-to-end data pipeline that automatically synchronizes, processes, and visualizes materials, manufacturing, and test data from all labs. Byterat also provides 24/7 access to experiments from anywhere in the world and integrates seamlessly with current workflows. It is customizable to specific cell chemistries and allows users to build custom visualizations, dashboards, and analyses. Byterat's AI-powered battery research has been published in leading journals, and its team has pioneered a new class of models that extract tell-tale signals of battery health from electrical signals to forecast future performance.
Tablesmith
Tablesmith is a free, privacy-first, and intuitive spreadsheet automation tool that allows users to build reusable data flows, effortlessly sort, filter, group, format, or split data across files/sheets based on cell values. It is designed to be easy to learn and use, with a focus on privacy and cross-platform compatibility. Tablesmith also offers an AI autofill feature that suggests and fills in information based on the user's prompt.
0 - Open Source Tools
9 - OpenAI Gpts
ImageJ Mentor
I assist biological image analysis, including ImageJ macro and Python coding.
Cell Tuber, Machine Assistant
Hello I'm Cell Tuber, Machine Assistant! What would you like help with today?
Stem Cell Regeneration Sage
Expert in biology, always ready to clarify new stem cell treatments.biomedical research, clinical trials. Learn about different stem cell types, current/future uses, and the latest in research.
Smartphone Repair Manual
A virtual smartphone repair manual offering detailed fixing instructions.

Repair Hero
Allround electronics repair expert with practical advise and video resources on range of devices. Think circular, avoid e-waste.
SCLC Atlas
Expert in SCLC research, focused on a specific paper and broader SCLC knowledge.